An interpretable machine learning model based on optimal feature selection for identifying CT abnormalities in patients with mild traumatic brain injury
- PMID: 40242564
- PMCID: PMC12002887
- DOI: 10.1016/j.eclinm.2025.103192
An interpretable machine learning model based on optimal feature selection for identifying CT abnormalities in patients with mild traumatic brain injury
Abstract
Background: Minor head trauma is a frequent cause of emergency department visits, early identification and prediction of mild traumatic brain injury (mTBI) patients with abnormal brain lesions are vital for minimizing unnecessary computed tomography (CT) scans, reducing radiation exposure, and ensuring timely effective treatment and care. This study aims to develop and validate an interpretable machine learning (ML) prediction model using routine laboratory data for guiding clinical decisions on CT scan use in mTBI patients.
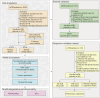
Methods: We conducted a multicentre study in China including data from January 2019 to July 2024. Our study included three patient cohorts: a retrospective training cohort (654 patients for training and 163 for internal testing) and two prospective validation cohorts (86 internal and 290 external patients). Fifty-one routine clinical laboratory characteristics, readily available from the electronic medical record (EMR) system within the first 24 h of admission, were collected. Seven ML algorithms were trained to develop predictive models, with the random forest (RF) algorithm used to optimize key feature combinations. Model predictive performance was evaluated using metrics such as the area under the receiver operating characteristic curve (AUC), positive predictive value (PPV), and F1 scores. The SHapley Additive exPlanation (SHAP) was applied to interpret the final model, while decision curve analysis (DCA) was used to assess the clinical net benefit.
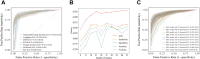
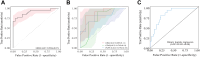
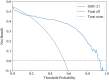
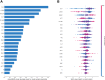
Findings: In the derivation cohort, 599 (73.3%) patients had normal CT scans and 218 (26.7%) had abnormal CT scans. The Gradient boosting classifier (GBC) model performed best among the seven ML models, with an AUC of 0.932 (95% CI: 0.900-0.963). After reducing features to 21 (8 biochemical test indicators, 3 coagulation markers, and 10 complete blood cell count indicators) according to feature importance rank, an explainable GBC-final model was established. The final model accurately predicted mTBI patients with abnormal CT in both internal (AUC 0.926, 95% CI: 0.893-0.958) and external (AUC 0.904, 95% CI: 0.835-0.973) validation cohorts. In the prospective cohort, final GBC model achieved AUC of 0.885 (95% CI: 0.753-1.000) and was significantly superior to traditional TBI biomarkers GFAP (AUC: 0.745) and PGP9.5 (AUC: 0.794). DCA revealed that the final model offered greater net benefits than "full intervention" or "no intervention" strategies within a probability threshold range of 0.16-0.93. SHAP analysis identified D-dimer levels, absolute lymphocyte and neutrophil counts, and hematocrit as key high-risk features.
Interpretation: Our optimal feature selection-based ML model accurately and reliably predicts CT abnormalities in mTBI patients using routine test data. By addressing clinicians' concerns regarding transparency and decision-making through SHAP and DCA analyses, we strengthen the potential clinical applicability of our ML model.
Funding: The Natural Science Foundation of Hubei Province, high-level Talent Research Startup Funding of Hubei University of Chinese Medicine, Wuhan Health and Family Planning Scientific Research Fund Project of Hubei Province, and Machine Learning-based Intelligent Diagnosis System for AFP-negative Liver Cancer Project.
Keywords: CT abnormal; DCA; Machine learning; Mild traumatic brain injury; Prediction model; SHAP.
© 2025 The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Jiang J.Y., Gao G.Y., Feng J.F., et al. Traumatic brain injury in China. Lancet Neurol. 2019;18(3):286–295. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

