Excessive activation?induced cytidine deaminase accumulated by proteasome inhibitors rescues abnormal class switch in activated B?cell?like diffuse large B?cell lymphoma
- PMID: 40242597
- PMCID: PMC12000863
- DOI: 10.3892/etm.2025.12863
Excessive activation?induced cytidine deaminase accumulated by proteasome inhibitors rescues abnormal class switch in activated B?cell?like diffuse large B?cell lymphoma
Abstract
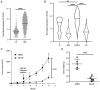
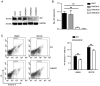
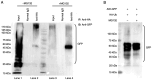
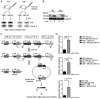
Activation-induced cytidine deaminase (AID) is an enzyme that plays a crucial role in mediating somatic hypermutation and class-switch recombination (CSR). It has been found to be associated with aberrant immunoglobulin CSR in activated B-cell-like diffuse large B-cell lymphoma (ABC-DLBCL). In the present study, MG132, a potent proteasome and calpain inhibitor, induced significant cell death in ABC-DLBCL cells and inhibited the growth of ABC-DLBCL cell xenograft tumors. The results also showed that MG132 induced AID accumulation by impairing proteasome degradation of AID. Excessive endogenous AID accumulation was observed in both AID-deficient and C57/BL6 wild-type mice treated with MG132, and apparent CSR of IgM to IgG1, IgG3 and IgE. Upon stimulation of cytokines such as LPS and/or IL-4, ABC-DLBCL cells also showed a noticeable increase in CSR of IgM to IgG1, IgG3 and IgE with decreased AID protein levels. The present study demonstrates that MG132 can induce AID accumulation, which in turn restores dysfunctional CSR in ABC-DLBCL. Using MG132 as a tool, the present study elucidates the anti-lymphoma effect of proteasome inhibitors on ABC-DLBCL by rescuing the abnormal AID-induced CSR.
Keywords: MG132; activated B-cell-like diffuse large B-cell lymphoma; activation-induced cytidine deaminase; class switch recombination; proteasome inhibitor; ubiquitination.
Copyright © 2025, Spandidos Publications.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma.J Exp Med. 2007 Mar 19;204(3):633-43. doi: 10.1084/jem.20062041. Epub 2007 Mar 12. J Exp Med. 2007. PMID: 17353367 Free PMC article.
-
A Hyper-IgM Syndrome Mutation in Activation-Induced Cytidine Deaminase Disrupts G-Quadruplex Binding and Genome-wide Chromatin Localization.Immunity. 2020 Nov 17;53(5):952-970.e11. doi: 10.1016/j.immuni.2020.10.003. Epub 2020 Oct 23. Immunity. 2020. PMID: 33098766 Free PMC article.
-
AID and TET2 co-operation modulates FANCA expression by active demethylation in diffuse large B cell lymphoma.Clin Exp Immunol. 2019 Feb;195(2):190-201. doi: 10.1111/cei.13227. Epub 2018 Nov 13. Clin Exp Immunol. 2019. PMID: 30357811 Free PMC article.
-
Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies.Adv Immunol. 2007;94:275-306. doi: 10.1016/S0065-2776(06)94009-7. Adv Immunol. 2007. PMID: 17560278 Review.
-
Regulation of Aicda expression and AID activity.Autoimmunity. 2013 Mar;46(2):83-101. doi: 10.3109/08916934.2012.749244. Epub 2013 Jan 17. Autoimmunity. 2013. PMID: 23181381 Free PMC article. Review.
References
-
- Crombie J. Classifying DLBCL subtypes for optimal treatment. Oncology (Williston Park) 2019;33(686504) - PubMed
-
- Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, Bhagat G, Borges AM, Boyer D, Calaminici M, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: Lymphoid neoplasms. Leukemia. 2022;36:1720–1748. doi: 10.1038/s41375-022-01620-2. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
