Exploring the role of EBV in multiple sclerosis pathogenesis through EBV interactome
- PMID: 40242760
- PMCID: PMC11999961
- DOI: 10.3389/fimmu.2025.1557483
Exploring the role of EBV in multiple sclerosis pathogenesis through EBV interactome
Abstract
Background: Epstein-Barr virus (EBV) is a known risk factor for multiple sclerosis (MS), even though the underlying molecular mechanisms are unclear and engage multiple immune pathways. Furthermore, the ultimate role of EBV in MS pathogenesis is still elusive. In contrast, Cytomegalovirus (CMV) has been identified as a protective factor for MS.
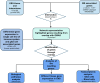
Objectives: This study aims to identify MS-associated genes that overlap with EBV interactome and to examine their expression in immune and glial cell subtypes.
Methods: We used P-HIPSTer, GWAS, and the Human Protein Atlas (HPA) to derive data on the EBV interactome, MS-associated genes and single-cell gene expression in immune and glial cells. The geneOverlap and dplyr R packages identified overlapping genes. A similar analysis was done for CMV and Adenovirus as negative control. Metascape and GTEx analyzed biological pathways and brain-level gene expression; transcriptomic analysis was performed on glial cells and peripheral blood in MS and controls. All the analyses performed in this study were generated using publicly available data sets.
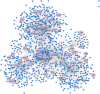
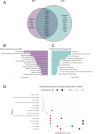
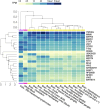
Results: We identified a "core" group of 21 genes shared across EBV interactome, MS genes, and immune and glial cells (p<0.001). Pathway analysis revealed expected associations, such as immune system activation, and unforeseen results, like the prolactin signaling pathway. BCL2 in astrocytes, MINK1 in microglia were significantly upregulated while AHI1 was downregulated in MS compared to controls.
Conclusions: Our findings offer novel insights into EBV and CMV interaction with immune and glial cells in MS, that may shed light on mechanisms involved in disease pathophysiology.
Keywords: CMV; EBV; MS pathogenesis; immune cells; interactome.
Copyright © 2025 Ballerini, Amoriello, Maghrebi, Bellucci, Addazio, Betti, Aprea, Masciulli, Caporali, Penati, Ballerini, De Meo, Portaccio, Salvetti and Amato.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Magliozzi R, Serafini B, Rosicarelli B, Chiappetta G, Veroni C, Reynolds R, et al. . B-cell enrichment and Epstein-Barr virus infection in inflammatory cortical lesions in secondary progressive multiple sclerosis. J Neuropathol Exp Neurol. (2013) 72:29–41. doi: 10.1097/NEN.0b013e31827bfc62 - DOI - PubMed
-
- Lassmann H, Niedobitek G, Aloisi F, Middeldorp JM, NeuroproMiSe EBV Working Group . Epstein-Barr virus in the multiple sclerosis brain: a controversial issue–report on a focused workshop held in the Centre for Brain Research of the Medical University of Vienna, Austria. Brain. (2011) 134:2772–86. doi: 10.1093/brain/awr197 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

