Transcriptome remodeling drives acclimation to iron availability in the marine N2-fixing cyanobacterium Trichodesmium erythraeum IMS101
- PMID: 40243322
- PMCID: PMC12090762
- DOI: 10.1128/msystems.01499-24
Transcriptome remodeling drives acclimation to iron availability in the marine N2-fixing cyanobacterium Trichodesmium erythraeum IMS101
Abstract
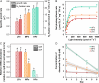
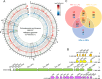
While enhanced phytoplankton growth as a result of iron (Fe) fertilization has been extensively characterized, our understanding of the underlying mechanisms remains incomplete. Here, we show in a laboratory setup mimicking Fe fertilization in the field that transcriptome remodeling is a primary driver of acclimation to Fe availability in the marine diazotrophic cyanobacterium Trichodesmium erythraeum IMS101. Fe supplementation promoted cell growth, photosynthesis and N2 fixation, and concomitant expression of the photosynthesis and N2 fixation genes. The expression of genes encoding major Fe-binding metalloproteins is tightly linked to cellular carbon and nitrogen metabolism and appears to be controlled by the ferric uptake regulator FurA, which is involved in regulating Fe uptake and homeostasis. This feedback loop is reinforced by substitutive expression of functionally equivalent or competitive genes depending on Fe availability, as well as co-expression of multiple Fe stress inducible isiA genes, an adaptive strategy evolved to elicit the Fe-responsive cascade. The study provides a genome-wide perspective on the acclimation of a prominent marine diazotroph to Fe availability, reveals an upgraded portfolio of indicator genes that can be used to better assess Fe status in the environment, and predicts scenarios of how marine diazotrophs may be affected in the future ocean.IMPORTANCEThe scarcity of trace metal iron (Fe) in global oceans has a great impact on phytoplankton growth. While enhanced primary productivity as a result of Fe fertilization has been extensively characterized, the underlying molecular mechanisms remain poorly understood. By subjecting the model marine diazotroph Trichodesmium erythraeum IMS101 to increasing concentrations of supplemented Fe, we demonstrate in it a comprehensively remodeled transcriptome that drives the mobilization of cellular Fe for coordinated carbon and nitrogen metabolism and reallocation of energy and resources. Our data provide broad genomic insight into marine diazotrophs acclimation to Fe availability, enabling the versatility and flexibility in choice of indicator genes for monitoring Fe status in the environment and having implications on how marine diazotrophs persist into the future ocean.
Keywords: Trichodesmium; iron limitation; isiA gene cluster; marine diazotroph; transcriptome remodeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Proteomic responses to ocean acidification of the marine diazotroph Trichodesmium under iron-replete and iron-limited conditions.Photosynth Res. 2019 Oct;142(1):17-34. doi: 10.1007/s11120-019-00643-8. Epub 2019 May 10. Photosynth Res. 2019. PMID: 31077001
-
Marine Colloids Boost Nitrogen Fixation in Trichodesmium erythraeum by Photoelectrophy.Environ Sci Technol. 2024 May 28;58(21):9236-9249. doi: 10.1021/acs.est.4c01849. Epub 2024 May 15. Environ Sci Technol. 2024. PMID: 38748855
-
Acclimation to medium-level non-lethal iron limitation: Adjustment of electron flow around the PSII and metalloprotein expression in Trichodesmium erythraeum IMS101.Biochim Biophys Acta Bioenerg. 2024 Jan 1;1865(1):149015. doi: 10.1016/j.bbabio.2023.149015. Epub 2023 Sep 23. Biochim Biophys Acta Bioenerg. 2024. PMID: 37742749
-
Interactions between CCM and N2 fixation in Trichodesmium.Photosynth Res. 2011 Sep;109(1-3):73-84. doi: 10.1007/s11120-010-9611-3. Epub 2010 Dec 29. Photosynth Res. 2011. PMID: 21190135 Review.
-
The nitrogen physiology of the marine N2-fixing cyanobacteria Trichodesmium spp.Trends Plant Sci. 2000 Apr;5(4):148-53. doi: 10.1016/s1360-1385(00)01576-4. Trends Plant Sci. 2000. PMID: 10740295 Review.
References
-
- Capone DG, Burns JA, Montoya JP, Subramaniam A, Mahaffey C, Gunderson T, Michaels AF, Carpenter EJ. 2005. Nitrogen fixation by Trichodesmium spp.: an important source of new nitrogen to the tropical and subtropical North Atlantic Ocean. Global Biogeochem Cycles 19:GB2024. doi:10.1029/2004GB002331 - DOI
-
- Capone DG, Zehr JP, Paerl HW, Bergman B, Carpenter EJ. 1997. Trichodesmium, a globally significant marine cyanobacterium. Science 276:1221–1229. doi:10.1126/science.276.5316.1221 - DOI
-
- Breitbarth E, Oschlies A, LaRoche J. 2007. Physiological constraints on the global distribution of Trichodesmium – effect of temperature on diazotrophy. Biogeosciences 4:53–61. doi:10.5194/bg-4-53-2007 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
