Impact of HIV-1 capsid polymorphisms on viral infectivity and susceptibility to lenacapavir
- PMID: 40243329
- PMCID: PMC12077089
- DOI: 10.1128/mbio.00187-25
Impact of HIV-1 capsid polymorphisms on viral infectivity and susceptibility to lenacapavir
Abstract
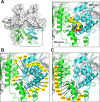
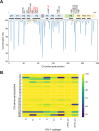
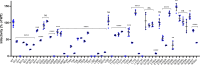
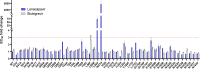
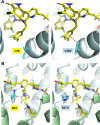
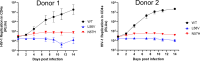
Lenacapavir (LEN) is a first-in-class capsid (CA) inhibitor for the treatment and prevention of HIV-1 infection. While LEN has shown potent antiviral activity across all major HIV-1 subtypes, the impact of existing HIV-1 CA sequence diversity on the activity of LEN remains to be determined. Here, we identified natural polymorphisms within the LEN-binding site and assessed each for their impact on viral infectivity and susceptibility to LEN. Using a co-crystal structure of LEN in complex with a CA hexamer, we identified 29 binding site residues within five angstroms of LEN and analyzed each for naturally occurring polymorphisms across a multiclade collection of >10,000 unique HIV-1 gag sequences. Eleven of these CA residues, including five (M66, Q67, K70, N74, and A105) previously associated with LEN resistance when mutated, were invariant across these sequences. The remaining 18 residues showed one or more substitutions with a ≥0.5% prevalence for a total of 54 CA polymorphisms. When introduced as site-directed mutants (SDMs) in an NL4.3-based reporter virus and evaluated for infectivity and drug susceptibility in MT-4 cells, 74% (40/54) showed impaired infectivity (0.01%-77% of wild type), with 96% (46/48) exhibiting minimal change (less than threefold) in susceptibility to LEN. While CA substitutions L56V and N57H conferred high-level resistance to LEN (72- and 4,890-fold, respectively), both variants showed diminished replication capacity in primary T-cells relative to the wild-type virus. Collectively, these results indicate that existing CA natural HIV-1 sequence diversity within the LEN-binding site is rare and should minimally impact LEN efficacy in treatment-naïve individuals.IMPORTANCEHIV-1 capsid protein mediates multiple essential functions throughout the viral replication cycle, making it an attractive target for therapeutic intervention. Lenacapavir (LEN), a first-in-class HIV-1 capsid inhibitor, is being evaluated as a long-acting option in multiple ongoing clinical studies for HIV treatment and prevention. Twice-yearly lenacapavir is approved in multiple countries for the treatment of adults with multi-drug-resistant HIV-1 in combination with other antiretrovirals, and its investigational use for pre-exposure prophylaxis has shown 99.9%-100% efficacy in preventing HIV infection among a broad and geographically diverse range of study participants. In this report, we investigated how HIV-1 sequence diversity within the LEN binding site may impact virus replication capacity and sensitivity to LEN. Our data demonstrate high capsid sequence conservation across a large and diverse collection of HIV-1 variants, with the majority of naturally occurring capsid polymorphisms having a detrimental effect on viral infectivity and minimal impact on susceptibility to LEN.
Keywords: HIV; antiretroviral resistance; lenacapavir; polymorphism; viral fitness.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- World Health Organization (WHO) . 2024. HIV and AIDS. Available from: https://www.who.int/news-room/fact-sheets/detail/hiv-aids. Retrieved 06 Jan 2025.
-
- Llibre JM, Clotet B. 2012. Once-daily single-tablet regimens: a long and winding road to excellence in antiretroviral treatment. AIDS Rev 14:168–178. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

