Lactococcus petauri LZys1 modulates gut microbiota, diminishes ileal FXR-FGF15 signaling, and regulates hepatic function
- PMID: 40243350
- PMCID: PMC12131734
- DOI: 10.1128/spectrum.01716-24
Lactococcus petauri LZys1 modulates gut microbiota, diminishes ileal FXR-FGF15 signaling, and regulates hepatic function
Abstract
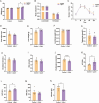
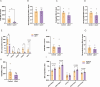
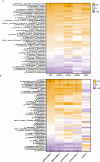
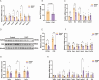
Recent studies have indicated that Lactococcus petauri LZys1 (L. petauri LZys1), isolated from healthy human feces, exhibits a promising probiotic profile in vitro. However, its impact on the physiological status of the host in vivo remains uncertain. The objective of our study was to investigate the effects and mechanisms of orally administering L. petauri LZys1 on gut microbiota and liver function in mice. We administered L. petauri LZys1 through daily oral gavage to C57BL/6 male mice. Subsequently, we analyzed changes in gut microbiota composition using 16S rRNA sequencing and quantified alterations in hepatic-intestinal bile acid (BA) profile. Serum biochemical parameters were assessed to evaluate liver function. Our findings revealed that L. petauri LZys1 led to an increase in body weight, liver mass, and serum aminotransferase levels. Oral administration altered the gut microbiota composition, resulting in reduced diversity and abundance of intestinal bacteria. Additionally, the profiles of BAs were suppressed across organs, associated with the downregulation of the ileum's farnesoid X receptor (FXR)/fibroblast growth factor 15 (FGF15) signaling pathway. The decrease in circulating FGF15 mediated the downregulation of hepatic fibroblast growth factor receptor 4 (FGFR4)/FXR, disrupting BA metabolism and fatty acid oxidation. Our findings suggest that L. petauri LZys1 may impact liver function by influencing the gut microbiota-mediated ileal FXR-FGF15 axis and inhibiting hepatic bile acid metabolism.
Importance: This work elucidated the impact of L. petauri LZys1 on host gut microbiota metabolism and hepatic physiological metabolism. We observed that L. petauri LZys1 administration induced liver weight gain and biochemical parameters changes, in addition to a altered gut microbiota and suppressed bile acid (BA) profiles. Furthermore, we propose that changes in liver status are related to the enterohepatic farnesoid X receptor-fibroblast growth factor axis, which alters bile acid metabolism and disrupts liver function. The above findings suggest that attention should be paid to the effect of probiotics on liver function.
Keywords: FGF15; FXR; Lactococcus petauri LZys1; gut microbiota; liver function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Inhibition of Farnesoid-x-receptor signaling during abdominal sepsis by dysbiosis exacerbates gut barrier dysfunction.Cell Commun Signal. 2025 May 21;23(1):236. doi: 10.1186/s12964-025-02224-w. Cell Commun Signal. 2025. PMID: 40399878 Free PMC article.
-
Sanye tablet regulates gut microbiota and bile acid metabolism to attenuate hepatic steatosis.J Ethnopharmacol. 2025 Apr 9;345:119514. doi: 10.1016/j.jep.2025.119514. Epub 2025 Feb 17. J Ethnopharmacol. 2025. PMID: 39971018
-
Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice.Cell Rep. 2014 Apr 10;7(1):12-8. doi: 10.1016/j.celrep.2014.02.032. Epub 2014 Mar 20. Cell Rep. 2014. PMID: 24656817
-
Molecular Basis of Bile Acid-FXR-FGF15/19 Signaling Axis.Int J Mol Sci. 2022 May 27;23(11):6046. doi: 10.3390/ijms23116046. Int J Mol Sci. 2022. PMID: 35682726 Free PMC article. Review.
-
Recent Advances in the Digestive, Metabolic and Therapeutic Effects of Farnesoid X Receptor and Fibroblast Growth Factor 19: From Cholesterol to Bile Acid Signaling.Nutrients. 2022 Nov 22;14(23):4950. doi: 10.3390/nu14234950. Nutrients. 2022. PMID: 36500979 Free PMC article. Review.
Cited by
-
Gut microbiota from Mori fructus (Morus alba L.) polyphenols and polysaccharides-dosed mice activates the PPARα/PGC-1α signaling pathway to mitigate HFD-induced metabolic syndrome in mice.Sci Rep. 2025 Aug 1;15(1):28137. doi: 10.1038/s41598-025-13715-8. Sci Rep. 2025. PMID: 40750814 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

