Genomic and phenotypic characterization of multidrug-resistant Staphylococcus haemolyticus isolated from burn patients in Chongqing, southwestern China
- PMID: 40243352
- PMCID: PMC12131843
- DOI: 10.1128/spectrum.02577-24
Genomic and phenotypic characterization of multidrug-resistant Staphylococcus haemolyticus isolated from burn patients in Chongqing, southwestern China
Abstract
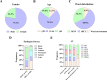
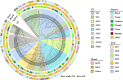
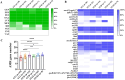
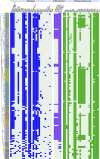
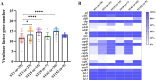
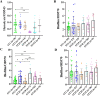
Staphylococcus hemolyticus is one of the frequently isolated nosocomial pathogens in immunocompromised patients; however, its genotypic and phenotypic characteristics in burn patients remain largely unknown. In this study, 146 S. haemolyticus strains were collected from the burn unit of a tertiary hospital in Chongqing, southwestern China. Whole genome sequencing was applied to determine the molecular characteristics of these S. haemolyticus isolates. Results showed that sequence type 1 (ST1; 26.7%, 39/146) was the most prevalent lineage, followed by clones ST42 (17.8%, 25/146), ST3 (15.8%, 23/146), ST25 (9.6%, 14/146), ST29 (4.8%, 7/146), and newly emerged ST152 (6.2%, 9/146). In terms of phenotypes, 90.4% (132/146) of the S. haemolyticus isolates were multidrug-resistant (MDR), and more than 90% strains exhibited resistance to penicillin, erythromycin, oxacillin, and levofloxacin. A total of 145 out of 146 S. haemolyticus isolates harbored at least 10 virulence factor genes. The tested strains with ST1, ST3, ST25, and ST42 presented similar hemolytic activities and biofilm formation capabilities. All detected ST152 isolates were MDR, corresponding to the number of their antimicrobial resistance genes being the largest among all tested STs. Moreover, ST152 isolates had increased hemolytic capacity and biofilm formation capability compared with their counterpart ST29 strains, the parent lineage of ST152 with a single locus variation. Overall, this study showed the genomic and phenotypic characteristics of 146 S. haemolyticus strains isolated in Chongqing and found the newly emerged ST152 MDR lineage with hypervirulence, which may call attention to the control of infections caused by MDR S. haemolyticus strains in hospitals.IMPORTANCEStaphylococcus haemolyticus is a common opportunistic pathogen with multidrug resistance in clinical infections. In this study, we analyzed the molecular epidemiological characteristics of 146 S. haemolyticus strains isolated from the burn patients of a tertiary hospital in Chongqing between 2017 and 2023. The results demonstrated that the phylogenetic evolution of S. haemolyticus strains was diverse and plastic. The majority of the isolates were multidrug-resistant (MDR). The virulence genes, resistance elements, and phenotypes, such as hemolysis and biofilm formation, were determined. A new lineage (ST152) that harbored the highest number of resistance genes was characterized to be 100% (9/9) MDR. The prevalence of ST152 S. haemolyticus strains suggests a new health threat in terms of control and treatment of coagulase-negative staphylococcal infections in Chongqing. This study provides information for clinical control of S. haemolyticus dissemination and infection in hospitals.
Keywords: ST152; Staphylococcus haemolyticus; antimicrobial resistance; biofilm formation; burn unit; hemolytic activity; virulence; whole genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Magnan C, Morsli M, Salipante F, Thiry B, Attar JE, Maio MD, Safaria M, Tran TA, Dunyach-Remy C, Ory J, Richaud-Morel B, Sotto A, Pantel A, Lavigne JP. 2024. Emergence of multidrug-resistant Staphylococcus haemolyticus in neonatal intensive care unit in Southern France, a genomic study. Emerg Microbes Infect 13:2353291. doi: 10.1080/22221751.2024.2353291 - DOI - PMC - PubMed
-
- Asante J, Hetsa BA, Amoako DG, Abia ALK, Bester LA, Essack SY. 2021. Multidrug-resistant coagulase-negative staphylococci isolated from bloodstream in the uMgungundlovu District of KwaZulu-Natal Province in South Africa: emerging pathogens. Antibiotics (Basel) 10:198. doi: 10.3390/antibiotics10020198 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

