Universal Prime Editing Therapeutic Strategy for RyR1-Related Myopathies: A Protective Mutation Rescues Leaky RyR1 Channel
- PMID: 40243436
- PMCID: PMC11988564
- DOI: 10.3390/ijms26072835
Universal Prime Editing Therapeutic Strategy for RyR1-Related Myopathies: A Protective Mutation Rescues Leaky RyR1 Channel
Abstract
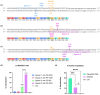
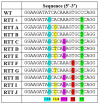
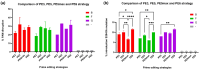
RyR1-related myopathies (RyR1-RMs) include a wide range of genetic disorders that result from mutations in the RYR1 gene. Pathogenic variants lead to defective intracellular calcium homeostasis and muscle dysfunction. Fixing intracellular calcium leaks by stabilizing the RyR1 calcium channel has been identified as a promising therapeutic target. Gene therapy via prime editing also holds great promise as it can cure diseases by correcting genetic mutations. However, as more than 700 variants have been identified in the RYR1 gene, a universal treatment would be a more suitable solution for patients. Our investigation into the RyR1-S2843A mutation has yielded promising results. Using a calcium leak assay, we determined that the S2843A mutation was protective when combined with pathogenic mutations and significantly reduced the Ca2+ leak of the RyR1 channel. Our study demonstrated that prime editing can efficiently introduce the protective S2843A mutation. In vitro experiments using the RNA electroporation of the prime editing components in human myoblasts achieved a 31% introduction of this mutation. This article lays the foundation for a new therapeutic approach for RyR1-RM, where a unique once-in-a-lifetime prime editing treatment could potentially be universally applied to all patients with a leaky RyR1 channel.
Keywords: CRISPR/Cas9; RYR1 gene; RyR1-RM; calcium channel; calcium leak; prime editing; protective mutation.
Conflict of interest statement
Columbia University and A.R.M. own stock in ARMGO, a company developing compounds targeting RyR, and have patents on Rycals. The remaining authors declare no competing interests. The authors have submitted a provisional patent application related to the work described in this manuscript.
Figures
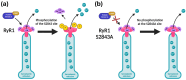



References
-
- Lawal T.A., Todd J.J., Witherspoon J.W., Bonnemann C.G., Dowling J.J., Hamilton S.L., Meilleur K.G., Dirksen R.T. Ryanodine receptor 1-related disorders: An historical perspective and proposal for a unified nomenclature. Skelet. Muscle. 2020;10:32. doi: 10.1186/s13395-020-00243-4. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

