The Impact of Soybean Genotypes on Rhizosphere Microbial Dynamics and Nodulation Efficiency
- PMID: 40243474
- PMCID: PMC11988558
- DOI: 10.3390/ijms26072878
The Impact of Soybean Genotypes on Rhizosphere Microbial Dynamics and Nodulation Efficiency
Abstract
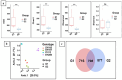
Rhizosphere microbiome exerts a significant role in plant health, influencing nutrient availability, disease resistance, and overall plant growth. Establishing a robust and efficient nodulation process is essential for optimal nitrogen fixation in legumes like soybeans. Different soybean genotypes exhibit variations in their rhizosphere microbiome, potentially impacting nitrogen fixation through nodulation. However, a detailed understanding of how specific soybean genotypes influence rhizosphere microbial communities and nodulation patterns remains limited. Our study aims to investigate the relationship between rhizosphere microbial abundance and plant growth in four soybean genotypes. We evaluated plant growth parameters, including biomass, leaf area, and stomatal conductance, and identified significant genotypic differences in nodulation. Specifically, genotypes PI 458505 and PI 603490 exhibited high levels of nodulation, while PI 605839A and PI 548400 displayed low nodulation. 16S rRNA gene amplicon sequencing revealed diverse bacterial communities in the rhizosphere, with Proteobacteria as the dominant phylum. High-nodulation genotypes harbored more diverse microbial communities enriched with Actinobacteria and Acidobacteriota, while low-nodulation genotypes showed higher abundances of Firmicutes and Planctomycetota. Alpha and beta diversity analyses confirmed distinct microbial community structures between high- and low-nodulation groups. Our findings suggest that the rhizosphere microbiome significantly influences soybean growth and nodulation, highlighting the potential for genotype-driven strategies to enhance plant-microbe interactions and improve soybean productivity.
Keywords: alpha diversity; amplicon sequencing; beta diversity; microbiome; stomata; taxonomic abundance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

