Bypassing Evolution of Bacterial Resistance to Phages: The Example of Hyper-Aggressive Phage 0524phi7-1
- PMID: 40243527
- PMCID: PMC11988461
- DOI: 10.3390/ijms26072914
Bypassing Evolution of Bacterial Resistance to Phages: The Example of Hyper-Aggressive Phage 0524phi7-1
Abstract
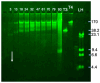
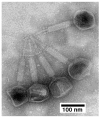
The ideal bacteriophages (phages) for the treatment of bacterial disease (phage therapy) would bypass bacterial evolution to phage resistance. However, this feature (called a hyper-aggression feature) has never been observed to our knowledge. Here, we microbiologically characterize, fractionate, genomically classify, and perform electron microscopy of the newly isolated Bacillus thuringiensis phage 0524phi7-1, which we find to have this hyper-aggression feature. Even visible bacterial colonies are cleared. Phage 0524phi7-1 also has three other features classified under hyper-aggression (four-feature-hyper-aggressive phage). (1) Phage 0524phi7-1 forms plaques that, although sometimes beginning as semi-turbid, eventually clear. (2) Clear plaques continue to enlarge for days. No phage-resistant bacteria are detected in cleared zones. (3) Plaques sometimes have smaller satellite plaques, even in gels so concentrated that the implied satellite-generating phage motion is not bacterial host generated. In addition, electron microscopy reveals that phage 0524phi7-1 (1) is a myophage with an isometric, 91 nm-head (diameter) and 210 nm-long contractile tail, and (2) undergoes extensive aggregation, which inhibits typical studies of phage physiology. The genome is linear double-stranded DNA, which, by sequencing, is 157.103 Kb long: family, Herelleviridae; genus, tsarbombavirus. The data suggest the hypothesis that phage 0524phi7-1 undergoes both swimming and hibernation. Techniques are implied for isolating better phages for phage therapy.
Keywords: bacteriophage, aggregation of; bacteriophage, hibernation and swimming of; bacteriophage, plaque morphology of; bacteriophage, screening of; electron microscopy; ultracentrifugation.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures







Similar articles
-
Isolation of A Novel Bacillus thuringiensis Phage Representing A New Phage Lineage and Characterization of Its Endolysin.Viruses. 2018 Nov 6;10(11):611. doi: 10.3390/v10110611. Viruses. 2018. PMID: 30404215 Free PMC article.
-
Genomic characterization and comparison of seven Myoviridae bacteriophage infecting Bacillus thuringiensis.Virology. 2016 Feb;489:243-51. doi: 10.1016/j.virol.2015.12.012. Epub 2016 Jan 14. Virology. 2016. PMID: 26773385
-
Existence of lysogenic bacteriophages in Bacillus thuringiensis type strains.J Invertebr Pathol. 2013 Jul;113(3):228-31. doi: 10.1016/j.jip.2013.04.008. Epub 2013 Apr 28. J Invertebr Pathol. 2013. PMID: 23632013
-
In-Gel Isolation and Characterization of Large (and Other) Phages.Viruses. 2020 Apr 7;12(4):410. doi: 10.3390/v12040410. Viruses. 2020. PMID: 32272774 Free PMC article. Review.
-
Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future.Viruses. 2014 Jul 9;6(7):2623-72. doi: 10.3390/v6072623. Viruses. 2014. PMID: 25010767 Free PMC article. Review.
References
-
- CDC (United States Centers for Disease Control and Prevention) Antibiotic Resistance Threats in the United States, 2019. Department of Health and Human Services, CDC; Atlanta, GA, USA: 2019. [(accessed on 21 July 2024)]. pp. 1–118. 2019 AR Threats Report. Available online: https://www.cdc.gov/antimicrobial-resistance/media/pdfs/2019-ar-threats-....
-
- Ahmad I., Malak H.A., Abulreesh H.H. Environmental antimicrobial resistance and its drivers: Apotential threat to public health. J. Glob. Antimicrob. Resist. 2021;27:101–111. - PubMed
-
- World Health Organization Antimicrobial Resistance. 2024. [(accessed on 21 July 2024)]. Available online: https://www.who.int/health-topics/antimicrobial-resistance.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

