The Arg108Cys Variant of Methylmalonyl-CoA Mutase: Clinical Implications for the Mexican Population Based on Molecular Dynamics and Docking
- PMID: 40243530
- PMCID: PMC11988910
- DOI: 10.3390/ijms26072887
The Arg108Cys Variant of Methylmalonyl-CoA Mutase: Clinical Implications for the Mexican Population Based on Molecular Dynamics and Docking
Abstract
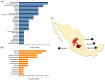
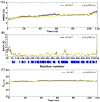
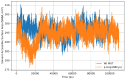
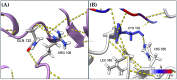
Methylmalonic acidemia (MMA) is a genetic condition associated with intellectual disability and a high mortality rate. It is caused by pathogenic variants in the MMUT gene, which codes methylmalonyl-CoA mutase enzyme (MUT). In the Mexican population, the variant NM_000255.4:c.322C>T or p.(Arg108Cys) is the most frequently found, but its structural pathogenic effect is scarcely studied. To describe the clinical picture of p.(Arg108Cys) homozygous patients and to predict its structural pathogenic effect, we performed an analysis of the medical files from six MMA Mexican p.(Arg108Cys) homozygous patients. The structural changes in MUT caused by this variant were analyzed through molecular dynamics simulations (MDS) and docking and compared with the wild-type (Wt) enzyme. The main clinical symptoms presented by the patients were feeding difficulties, lethargy, and neurodevelopmental delay, with a predominance of early-onset phenotype and a mortality rate of 83%. We found significant structural changes in MUT structure, particularly in the catalytic domain, with increased volume cavity, shortening of the binding substrate tunnel, and aberrant accommodation. Also, the dimerization interface area increased from 1343 Å2 in the Wt to 3386 Å2, and the dimer formation involved a different set of amino acids. The NM_000255.4:c.322C>T or p.(Arg108Cys) MMUT variant is associated with a severe outcome in MMA Mexican patients, and the enzyme was associated with ostentatious topological changes in the secondary and tertiary structure, which impacted the catalytic domain, the accommodation of the substrate, and the dimerization interface. Further ex vivo functional studies are needed to confirm these predictions, such as enzymatic activity measurements in fibroblasts of patients.
Keywords: genetic diseases; inborn errors of metabolism; methylmalonic acidemia; propionate defects; rare diseases.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Fagerberg L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014;13:397–406. - PMC - PubMed
-
- Frenkel E.P., Kitchens R.L. Intracellular Localization of Hepatic Propionyl-CoA Carboxylase and Methylmalonyl-CoA Mutase in Humans and Normal and Vitamin B12 Deficient Rats. Br. J. Haematol. 1975;31:501–513. - PubMed
-
- Acquaviva C., Benoist J.F., Pereira S., Callebaut I., Koskas T., Porquet D., Elion J. Molecular basis of methylmalonyl-CoA mutase apoenzyme defect in 40 European patients affected by mut° and mut–forms of methylmalonic acidemia: Identification of 29 novel mutations in the MUT gene. Hum. Mutat. 2005;25:167–176. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

