Subgenomic RNA and Limited Cross-Reactive Neutralising Antibodies Point to Potential Improvements in SARS-CoV-2 Clinical Handling
- PMID: 40243564
- PMCID: PMC11988571
- DOI: 10.3390/ijms26072948
Subgenomic RNA and Limited Cross-Reactive Neutralising Antibodies Point to Potential Improvements in SARS-CoV-2 Clinical Handling
Abstract
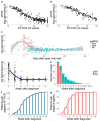
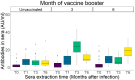
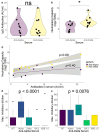
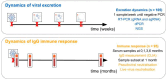
The current clinical management of SARS-CoV-2 disease control and immunity may be not optimal anymore. Reverse transcription polymerase chain reaction (RT-PCR) of genomic viral RNA is broadly used for diagnosis, even though the virus may still be detectable when it is already non-infectious. Regarding serology, commercial assays mostly still rely on ancestral spike detection despite significant changes in the genetic sequence of the current circulating variants. We followed a group of 105 non-vaccinated individuals, measuring their viral shedding until negativity and antibody response up to six months. The mean viral detection period until a negative RT-PCR result was 2.2 weeks when using subgenomic RNA-E as a detection target, and 5.2 weeks when using genomic RNA as a detection target. Our neutralising antibody results suggest that, when challenged against a variant different from the variant of first exposure, commercial immunoassays are suboptimal at predicting the neutralising capacity of sera. Additionally, anti-Alpha and anti-Delta antibodies showed very low cross-reactivity between variants. This study provides insights into viral shedding and immune response in pre-Omicron variants like Alpha and Delta, which have been understudied in the published literature. These conclusions point to potential improvements in the clinical management of SARS-CoV-2 cases in order to organise vaccination campaigns and select monoclonal antibody treatments.
Keywords: SARS-CoV-2; neutralising antibodies; subgenomic RNA; viral shedding dynamics.
Conflict of interest statement
Authors Almudena Rojas and Joaquin Mendoza are from VIRCELL S.L, the authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
-
- Richard-Greenblatt M., Ziegler M.J., Bromberg V., Huang E., Abdallah H., Tolomeo P., Lautenbach E., Glaser L., Kell B.J. Quantifying the Impact of Nasopharyngeal Specimen Quality on Severe Acute Respiratory Syndrome Coronavirus 2 Test Performance. Open Forum Infect. Dis. 2021;8:ofab235. doi: 10.1093/ofid/ofab235. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

