Cell Wall-Based Machine Learning Models to Predict Plant Growth Using Onion Epidermis
- PMID: 40243585
- PMCID: PMC11989001
- DOI: 10.3390/ijms26072946
Cell Wall-Based Machine Learning Models to Predict Plant Growth Using Onion Epidermis
Abstract
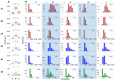
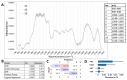
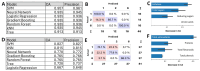
The plant cell wall (CW) is a physical barrier that plays a dual role in plant physiology, providing structural support for growth and development. Understanding the dynamics of CW growth is crucial for optimizing crop yields. In this study, we employed onion (Allium cepa L.) epidermis as a model system, leveraging its layered organization to investigate growth stages. Microscopic analysis revealed proportional variations in cell size in different epidermal layers, offering insights into growth dynamics and CW structural adaptations. Fourier transform infrared spectroscopy (FTIR) identified 11 distinct spectral intervals associated with CW components, highlighting structural modifications that influence wall elasticity and rigidity. Biochemical assays across developmental layers demonstrated variations in cellulose, soluble sugars, and antioxidant content, reflecting biochemical shifts during growth. The differential expression of ten cell wall enzyme (CWE) genes, analyzed via RT-qPCR, revealed significant correlations between gene expression patterns and CW composition changes across developmental layers. Notably, the gene expression levels of the pectin methylesterase and fucosidase enzymes were associated with the contents in cellulose, soluble sugar, and antioxidants. To complement these findings, machine learning models, including Support Vector Machines (SVM), k-Nearest Neighbors (kNN), and Neural Networks, were employed to integrate FTIR data, biochemical parameters, and CWE gene expression profiles. Our models achieved high accuracy in predicting growth stages. This underscores the intricate interplay among CW composition, CW enzymatic activity, and growth dynamics, providing a predictive framework with applications in enhancing crop productivity and sustainability.
Keywords: Allium cepa L.; cell wall composition; cell wall enzymes; machine learning; modeling; onion epidermis; plant growth.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
MeSH terms
Substances
Grants and funding
- Convenio Plurianual con la Universidad Politécnica de Madrid en la línea de actuación Programa de Excelencia para el Profesorado Universitario./Comunidad de Madrid
- VAGI23JPMC/Universidad Politécnica de Madrid
- VAGI24JPMC/Universidad Politécnica de Madrid
- 057Bis/PG/Espagne/2020-2021/Algerian government
- ChaSisCOMA (PID2021-122711NB-C21)/Ministerio de Ciencia, Innovación y Universidades
LinkOut - more resources
Full Text Sources

