Characterizing Common Factors Affecting Replication Initiation During H2O2 Exposure and Genetic Mutation-Induced Oxidative Stress in Escherichia coli
- PMID: 40243598
- PMCID: PMC11989076
- DOI: 10.3390/ijms26072968
Characterizing Common Factors Affecting Replication Initiation During H2O2 Exposure and Genetic Mutation-Induced Oxidative Stress in Escherichia coli
Abstract
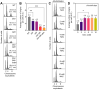
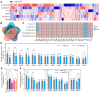
Oxidative stress is prevalent in organisms, and excessive oxidative damage can trigger cell death. Bacteria have evolved multiple pathways to cope with adverse stress, including the regulation of the cell cycle. Previous studies show that non-lethal exposure to H2O2 and mutations in antioxidant enzymes suppress replication initiation in Escherichia coli. The existence of common regulatory factors governing replication initiation across diverse causes-induced oxidative stress remains unclear. In this study, we utilized flow cytometry to determine the replication pattern of E. coli, and found that oxidative stress also participated in the inhibition of replication initiation by a defective iron regulation (fur-bfr-dps deletion). Adding a certain level of ATP promoted replication initiation in various antioxidant enzyme-deficient mutants and the ΔfurΔbfrΔdps mutant, suggesting that low ATP levels could be a common factor in the inhibition of replication initiation by different causes-induced oxidative stress. More potential common factors were screened using proteomics, followed by genetic validation with H2O2 stress. We found that oxidative stress might mediate the inhibition of replication initiation by interfering with the metabolism of glycine, glutamate, ornithine, and aspartate. Blocking CcmA-dependent cytochrome c biosynthesis, deleting the efflux pump proteins MdtABCD and TolC, or the arabinose transporter AraFHG eliminated the replication initiation inhibition by H2O2. In conclusion, this study uncovers a common multifactorial pathway of different causes-induced oxidative stress inhibiting replication initiation. Dormant and persistent bacteria exhibit an arrested or slow cell cycle, and non-lethal oxidative stress promotes their formation. Our findings contribute to exploring strategies to limit dormant and persistent bacterial formation by maintaining faster DNA replication initiation (cell cycle progression).
Keywords: ATP levels; DNA replication initiation; efflux pumps; metabolism; oxidative stress.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

