DNA Methylation Patterns Provide Insights into the Epigenetic Regulation of Intersex Formation in the Chinese Mitten Crab (Eriocheir sinensis)
- PMID: 40244073
- PMCID: PMC11989155
- DOI: 10.3390/ijms26073224
DNA Methylation Patterns Provide Insights into the Epigenetic Regulation of Intersex Formation in the Chinese Mitten Crab (Eriocheir sinensis)
Abstract
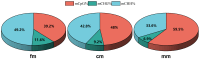
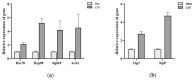
DNA methylation is a form of epigenetic regulation that plays an important role in regulating gene expression of organisms. However, the DNA methylation pattern of intersex crabs has not yet been clarified. In order to reveal the DNA methylation in intersex Eriocheir sinensis, this study investigated the genome-wide DNA methylation profiles of female, male, and intersex individuals. The similar results across samples showed that the levels of cytosine methylation in the CG context were significantly higher than that in the CHG and CHH contexts. The methylation levels in the promoter region were higher than those in other functional element regions. We screened 149 differentially methylated genes (DMGs) in the promoter region between female and intersex crabs and 110 DMGs between male and intersex crabs. Three core gene networks were found in a comparison group of female and intersex crabs that involved heat shock proteins, ribosomes, and metabolism pathways; two core gene networks were found in the comparison group of male and intersex crabs that involved ribosomes and metabolism pathways. The six confirmed genes of Hsc70, Hsp90, Rpl18, Acsl1, Yip2, and Rpl7 had lower methylation levels in the promoter region of intersex crabs than that of female and male crabs. However, six genes showed higher expression in intersex crabs than in female and male crabs. Our results reveal that DNA methylation is involved in the formation and maintenance of life activities of intersex crabs through the regulation of gene expression, enriching the DNA methylation library of the whole genome of E. sinensis and providing new insights for a better understanding of the epigenetic regulation of the formation of intersex E. sinensis.
Keywords: DNA methylation; Eriocheir sinensis; intersex.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Allis C.D., Jenuwein T. The Molecular Hallmarks of Epigenetic Control. Nat. Rev. Genet. 2016;17:487–500. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

