The Role of Killer Ig-like Receptors in Diseases from A to Z
- PMID: 40244151
- PMCID: PMC11989319
- DOI: 10.3390/ijms26073242
The Role of Killer Ig-like Receptors in Diseases from A to Z
Abstract
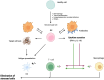
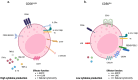
Killer Ig-like Receptors (KIRs) regulate immune responses, maintaining the balance between activation and inhibition of the immune system. KIRs are expressed on natural killer cells and some CD8 T cells and interact with HLA class I molecules, influencing various physiological and pathological processes. KIRs' polymorphism creates a variability in immune responses among individuals. KIRs are involved in autoimmune disorders, cancer, infections, neurological diseases, and other diseases. Specific combinations of KIRs and HLA are linked to several diseases' susceptibility, progression, and outcomes. In particular, the balance between inhibitory and activating KIRs can determine how the immune system responds to pathogens and tumors. An imbalance can lead to an excessive response, contributing to autoimmune diseases, or an inadequate response, allowing immune evasion by pathogens or cancer cells. The increasing number of studies on KIRs highlights their essential role as diagnostic and prognostic biomarkers and potential therapeutic targets. This review provides a comprehensive overview of the role of KIRs in all clinical conditions and diseases, listed alphabetically, where they are analyzed.
Keywords: KIR; autoimmune diseases; genetic; polymorphisms.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

