Advancing Personalized Medicine in Alzheimer's Disease: Liquid Biopsy Epigenomics Unveil APOE ε4-Linked Methylation Signatures
- PMID: 40244264
- PMCID: PMC11989983
- DOI: 10.3390/ijms26073419
Advancing Personalized Medicine in Alzheimer's Disease: Liquid Biopsy Epigenomics Unveil APOE ε4-Linked Methylation Signatures
Abstract
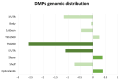
Recent studies show that patients with Alzheimer's disease (AD) harbor specific methylation marks in the brain that, if accessible, could be used as epigenetic biomarkers. Liquid biopsy enables the study of circulating cell-free DNA (cfDNA) fragments originated from dead cells, including neurons affected by neurodegenerative processes. Here, we isolated and epigenetically characterized plasma cfDNA from 35 patients with AD and 35 cognitively healthy controls by using the Infinium® MethylationEPIC BeadChip array. Bioinformatics analysis was performed to identify differential methylation positions (DMPs) and regions (DMRs), including APOE ε4 genotype stratified analysis. Plasma pTau181 (Simoa) and cerebrospinal fluid (CSF) core biomarkers (Fujirebio) were also measured and correlated with differential methylation marks. Validation was performed with bisulfite pyrosequencing and bisulfite cloning sequencing. Epigenome-wide cfDNA analysis identified 102 DMPs associated with AD status. Most DMPs correlated with clinical cognitive and functional tests including 60% for Mini-Mental State Examination (MMSE) and 80% for Global Deterioration Scale (GDS), and with AD blood and CSF biomarkers. In silico functional analysis connected 30 DMPs to neurological processes, identifying key regulators such as SPTBN4 and APOE genes. Several DMRs were annotated to genes previously reported to harbor epigenetic brain changes in AD (HKR1, ZNF154, HOXA5, TRIM40, ATG16L2, ADAMST2) and were linked to APOE ε4 genotypes. Notably, a DMR in the HKR1 gene, previously shown to be hypermethylated in the AD hippocampus, was validated in cfDNA from an orthogonal perspective. These results support the feasibility of studying cfDNA to identify potential epigenetic biomarkers in AD. Thus, liquid biopsy could improve non-invasive AD diagnosis and aid personalized medicine by detecting epigenetic brain markers in blood.
Keywords: APOE ε4; Alzheimer’s disease; DNA methylation; EPIC array; blood; cell-free DNA; liquid biopsy.
Conflict of interest statement
D.A. participated in advisory boards from Fujirebio-Europe, Roche Diagnostics, Grifols S.A., and Lilly, and received speaker honoraria from Fujirebio-Europe, Roche Diagnostics, Nutricia, Krka Farmacéutica S.L., Zambon S.A.U., and Esteve Pharmaceuticals S.A. D.A. declares a filed patent application (WO2019175379 A1 markers of synaptopathy in neurodegenerative disease). The funders had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- Alzheimers Disease International World Alzheimer Report 2018. 2018. [(accessed on 2 April 2025)]. Available online: https://www.alz.co.uk/research/WorldAlzheimerReport2018.pdf.
-
- Alzheimers Disease International World Alzheimer Report 2021. 2021. [(accessed on 2 April 2025)]. Available online: https://www.alzint.org/u/World-Alzheimer-Report-2021.pdf.
-
- Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

