Fully Phased Population-Prevalent East African Cattle BoLA-I Alleles Determined Using PacBio HiFi Long-Read Sequencing Represent Five Novel Specificities With Distinctive Peptide Binding Potential
- PMID: 40244593
- PMCID: PMC12005355
- DOI: 10.1111/tan.70183
Fully Phased Population-Prevalent East African Cattle BoLA-I Alleles Determined Using PacBio HiFi Long-Read Sequencing Represent Five Novel Specificities With Distinctive Peptide Binding Potential
Abstract
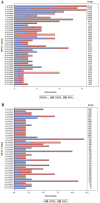
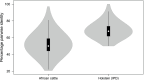
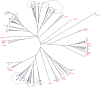
Due to factors such as lower biosecurity, greater wildlife/farm animal interfaces, and environmental challenges, cattle in sub-Saharan Africa are exposed to more diverse and intensive bacterial, viral and protozoan pathogen challenges than cattle in Europe and other high-income regions of the world. Classical class I genes of the major histocompatibility complex (MHC) contribute to protection from diseases caused by these pathogens by refining a huge pool of potential pathogen-derived peptide ligands into a smaller ensemble for presentation to CD8+ T cells. Knowledge of population-prevalent MHC alleles is therefore critical for evidence-based approaches to vaccine design and improved understanding of pathogen resistance. Whereas variation in MHC molecules is understood in most detail for European Bos taurus, the alleles expressed by Africa's cattle remain poorly defined. We have leveraged recent improvements in the accuracy of PacBio high-fidelity (HiFi) circular consensus sequencing (CCS) and adapted stringent sequence filtering algorithms to identify hundreds of as yet uncharacterised fully phased BoLA-I alleles from multiple populations of African taurine (Ankole) and indicine (Zebu) cattle in East Africa. The analysis highlights a convergence of population-prevalent class I MHC allelic repertoires in taurine and indicine cattle, likely due to the similar pathogen-driven selective pressures. Our analysis of the anchor residue accommodating pockets of these prevalent alleles revealed extremely high levels of polymorphism, which contrast with Holstein alleles that exhibit a more limited repertoire of MHC specificity-determining pocket residues, potentially constraining the breadth of peptide presentation. However, in the context of considerable sequence and physicochemical variation in the pocket-forming residues, it was possible to discern overlaps in the predicted peptide binding spectrum. Interrogation of potential differences in peptide binding specificities with European B. taurus alleles revealed that the fully phased African cattle class I MHC alleles represent five novel specificities. We envisage that this novel finding will find broad application in assessing potentially achievable vaccination coverages of future pathogen-encoded vaccine candidates against important intracellular pathogens. One aim of future research should be to leverage recent improvements in the sensitivity of mass spectrometry combined with immunoprecipitation of peptides bound to African cattle MHC to search directly for T-cell epitopes in the context of the inferred 'supertype' diversity.
Keywords: MHC; cattle; high‐throughput sequencing.
© 2025 The Author(s). HLA: Immune Response Genetics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Sequence diversity between class I MHC loci of African native and introduced Bos taurus cattle in Theileria parva endemic regions: in silico peptide binding prediction identifies distinct functional clusters.Immunogenetics. 2016 May;68(5):339-52. doi: 10.1007/s00251-016-0902-5. Epub 2016 Feb 6. Immunogenetics. 2016. PMID: 26852329
-
A comparative study of major histocompatibility complex antigens in east African and European cattle breeds.Anim Genet. 1988;19(1):17-29. doi: 10.1111/j.1365-2052.1988.tb00784.x. Anim Genet. 1988. PMID: 3377275
-
Characterization of binding specificities of bovine leucocyte class I molecules: impacts for rational epitope discovery.Immunogenetics. 2014 Dec;66(12):705-18. doi: 10.1007/s00251-014-0802-5. Epub 2014 Sep 4. Immunogenetics. 2014. PMID: 25186069 Free PMC article.
-
Genetic variation and responses to vaccines.Anim Health Res Rev. 2004 Dec;5(2):197-208. doi: 10.1079/ahr200469. Anim Health Res Rev. 2004. PMID: 15984325 Review.
-
Designing bovine T cell vaccines via reverse immunology.Ticks Tick Borne Dis. 2012 Jun;3(3):188-92. doi: 10.1016/j.ttbdis.2011.12.001. Epub 2012 Jan 9. Ticks Tick Borne Dis. 2012. PMID: 22621863 Review.
References
-
- Kelley J., Walter L., and Trowsdale J., “Comparative Genomics of Major Histocompatibility Complexes,” Immunogenetics 56, no. 10 (2005): 683–695. - PubMed
-
- Codner G. F., Stear M. J., Reeve R., Matthews L., and Ellis S. A., “Selective Forces Shaping Diversity in the Class I Region of the Major Histocompatibility Complex in Dairy Cattle,” Animal Genetics 43, no. 3 (2012): 239–249. - PubMed
-
- Epstein H., The Origin of the Domesticated Animals of Africa, vol. 1 (Africana Publishing Corporation, 1971), 1–573.
-
- Epstein H. and Mason L., “Cattle,” in Evolution of Domesticated Animals, ed. Mason I. L. (Longman Group, 1984).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

