Application of Metagenomic Long-Read Sequencing for the Diagnosis of Herpetic Uveitis
- PMID: 40244604
- PMCID: PMC12013679
- DOI: 10.1167/iovs.66.4.50
Application of Metagenomic Long-Read Sequencing for the Diagnosis of Herpetic Uveitis
Abstract
Purpose: To investigate the sensitivity and specificity of herpes virus detection by nanopore metagenomic analysis (NMA) compared with multiplex polymerase chain reaction (mPCR)-positive and -negative controls.
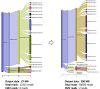
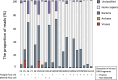
Methods: This study included 43 patients with uveitis who had been screened for intraocular herpes virus infection using mPCR from aqueous humor samples. Aqueous humor samples stored after mPCR were subjected to whole-genome amplification, long-read sequencing, and analysis of the phylogenetic microorganism composition using a Flongle flow cell on the Oxford Nanopore MinION platform. For samples that tested positive with mPCR and negative with the Flongle flow cell, additional long-read sequencing was performed using a MinION flow cell, which enabled acquisition of more sequence data. The sensitivity and specificity of herpes virus detection by NMA were compared with the mPCR-positive and -negative controls.
Results: NMA using a Flongle flow cell detected the pathogenic virus in 60.0% of those who tested positive by mPCR (12/20). Further analysis using the MinION flow cell successfully identified viral DNA fragments in three out of the eight initially undetected samples, yielding a collective sensitivity of 75.0% (15/20). All of the virus detected with the long-read sequencing were identical to those diagnosed by mPCR testing, and none of the samples that tested negative by mPCR revealed herpes viral DNA with the use of long-read sequencing.
Conclusions: For the detection of etiologic herpes virus DNA fragments, NMA revealed a reasonable sensitivity and high specificity. Our study highlights the potential of nanopore sequencing to facilitate further advances in uveitis diagnosis.
Conflict of interest statement
Disclosure:
Figures

References
-
- Sugita S, Takase H, Nakano S.. Practical use of multiplex and broad-range PCR in ophthalmology. Jpn J Ophthalmol. 2021; 65(2): 155–168. - PubMed
-
- Sugita S, Ogawa M, Shimizu N, et al.. Use of a comprehensive polymerase chain reaction system for diagnosis of ocular infectious diseases. Ophthalmology. 2013; 120(9): 1761–1768. - PubMed
-
- Nakano S, Tomaru Y, Kubota T, et al.. Multiplex solid-phase real-time polymerase chain reaction without DNA extraction: a rapid intraoperative diagnosis using microvolumes. Ophthalmology. 2021; 128(5): 729–739. - PubMed
-
- Sonoda KH, Hasegawa E, Namba K, et al.. Epidemiology of uveitis in Japan: a 2016 retrospective nationwide survey. Jpn J Ophthalmol. 2021; 65(2): 184–190. - PubMed
-
- Mercanti A, Parolini B, Bonora A, Lequaglie Q, Tomazzoli L.. Epidemiology of endogenous uveitis in north-eastern Italy. Analysis of 655 new cases. Acta Ophthalmol Scand. 2001; 79(1): 64–68. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

