RRM2B deficiency causes dATP and dGTP depletion through enhanced degradation and slower synthesis
- PMID: 40244665
- PMCID: PMC12037051
- DOI: 10.1073/pnas.2503531122
RRM2B deficiency causes dATP and dGTP depletion through enhanced degradation and slower synthesis
Abstract
Mitochondrial DNA (mtDNA) replication requires a steady supply of deoxyribonucleotides (dNTPs), synthesized de novo by ribonucleotide reductase (RNR). In nondividing cells, RNR consists of RRM1 and RRM2B subunits. Mutations in RRM2B cause mtDNA depletion syndrome, linked to muscle weakness, neurological decline, and early mortality. The impact of RRM2B deficiency on dNTP pools in nondividing tissues remains unclear. Using a mouse knockout model, we demonstrate that RRM2B deficiency selectively depletes dATP and dGTP, while dCTP and dTTP levels remain stable or increase. This depletion pattern resembles the effects of hydroxyurea, an inhibitor that reduces overall RNR activity. Mechanistically, we propose that the depletion of dATP and dGTP arises from their preferred degradation by the dNTPase SAMHD1 and the lower production rate of dATP by RNR. Identifying dATP and dGTP depletion as a hallmark of RRM2B deficiency provides insights for developing nucleoside bypass therapies to alleviate the effects of RRM2B mutations.
Keywords: dNTP metabolism; genome stability; mtDNA stability; ribonucleotide reductase.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
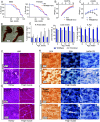
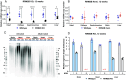


References
-
- Nordlund P., Reichard P., Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 (2006). - PubMed
-
- Bourdon A., et al. , Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 39, 776–780 (2007). - PubMed
-
- Keshavan N., et al. , The natural history of infantile mitochondrial DNA depletion syndrome due to RRM2B deficiency. Genet. Med. 22, 199–209 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

