Exploring the Role of T-Cell Metabolism in Modulating Immunotherapy Efficacy for Non-Small Cell Lung Cancer Based on Clustering
- PMID: 40244859
- PMCID: PMC12217653
- DOI: 10.1002/jcla.25020
Exploring the Role of T-Cell Metabolism in Modulating Immunotherapy Efficacy for Non-Small Cell Lung Cancer Based on Clustering
Abstract
Background: Immunotherapy, especially immune checkpoint blockade (ICB) therapy, has demonstrated noteworthy advancements in the realm of non-small cell lung cancer (NSCLC). However, the efficacy of ICB therapy is limited to a small subset of patients with NSCLC, and the underlying mechanisms remain poorly understood.
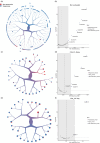
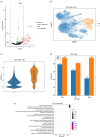
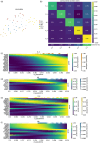
Study design and discoveries: In this study, we conducted a comprehensive investigation of the metabolic profiles of infiltrating T cells in NSCLC tumors and revealed the metabolic heterogeneity, which associated with the prognosis of ICB therapy, in three T-cell subtypes. After metabolic clustering, we split these metabolic clusters into two groups: Nonresponse-associated (NR) clusters that enriched with cells from nonresponders, and response-associated (R) clusters that not belonging to NR clusters. Then, we elucidated their metabolic differences and specific functions. Notably, we discovered HSPA1A was significantly downregulated in NR clusters of all three T-cell subtypes. In addition, leveraging single-cell T-cell receptor sequencing data and pseudotime series analysis, we revealed the reciprocal interconversion between R and NR metabolic clusters within the same T-cell clone. This suggests a potential metabolic reprogramming capability of T cells. Furthermore, through the analysis of intercellular communication, we identified the specific intercellular signaling in the R clusters, which might promote the activation and regulation of signal transduction pathways that affect the prognosis of ICB therapy.
Conclusion: In conclusion, our study offers substantial insights into the mechanisms of relationships between T-cell metabolisms and ICB therapy outcomes, shedding light on the mechanism of immunotherapy efficacy in patients with NSCLC. Such investigations will contribute to overcoming treatment resistance.
Keywords: T cell; clustering; immune metabolism; immunotherapy; non‐small cell lung cancer.
© 2024 The Authors. Journal of Clinical Laboratory Analysis published by Wiley Periodicals LLC.
Conflict of interest statement
Rui Hou and Yue Gong are employed by Genies Beijing Co., Ltd. All other authors declare no competing interests.
Figures

References
-
- Oliver A. L., “Lung Cancer: Epidemiology and Screening,” The Surgical Clinics of North America 102, no. 3 (2022): 335–344. - PubMed
-
- Hynes PR, Das MJ. Stereotactic Radiosurgery (SRS) and Stereotactic Body Radiotherapy (SBRT). 2023. Jul 25. In: StatPearls [Internet]. Treasure Island: (FL): StatPearls Publishing; 2024 Jan–. PMID: 31194323. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

