Transcriptome and network analysis pinpoint ABA and plastid ribosomal proteins as main contributors to salinity tolerance in the rice variety, CSR28
- PMID: 40244966
- PMCID: PMC12005493
- DOI: 10.1371/journal.pone.0321181
Transcriptome and network analysis pinpoint ABA and plastid ribosomal proteins as main contributors to salinity tolerance in the rice variety, CSR28
Abstract
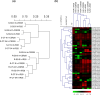
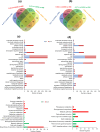
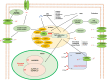
Salinity stress is a major challenge for rice production, especially at seedling stage. To gain comprehensive insight into the molecular mechanisms and potential candidate genes involved in rice salinity stress response, we integrated physiological, transcriptome and network analysis to investigate salinity tolerance in two contrasting rice genotypes. The root and shoot samples were collected at two timepoints (6 hours and 54 hours) of high salt treatment. Element assay showed that the tolerant genotype CSR28 had lower Na+/K+ ratio in both organs than in those of the sensitive genotype IR28 under salinity stress. A total of 15,483 differentially expressed genes (DEGs) were identified from the RNA-Seq analysis. The salt-specific genes were mainly involved in metabolic processes, response to stimulus, and transporter activity, and were enriched in key metabolic pathways such as, biosynthesis of secondary metabolites, plant hormone signal transduction, and carotenoid biosynthesis. Furthermore, the results showed that the differential genes involved in abscisic acid (ABA) biosynthesis were specifically up-regulated in the tolerant genotype. Network analysis revealed 50 hub genes for the salt-specific genes in the roots of CSR28 which mainly encodes ribosomal proteins (RPs). Functional validation of the nine hub genes revealed three plastid RPs (PRPs), including OsPRPL17, OsPRPS9 and OsPRPL11, which contributes to protein synthesis, chloroplast development and stress signaling. Our findings suggested that ABA and PRPs play key roles to enhance of salinity tolerance in CSR28. Our study provides valuable information for further investigations of the candidate genes associated with salt tolerance and the development of salt-tolerant rice varieties.
Copyright: © 2025 Akbarzadeh Lelekami et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Hunter MC, Smith RG, Schipanski ME, Atwood LW, Mortensen DA. Agriculture in 2050: Recalibrating targets for sustainable Intensification. BioScience. 2017;67(4):386–91. doi: 10.1093/biosci/bix010 - DOI
-
- Bazzaz F, Sombroek W. Global climatic change and agricultural production: An assessment of current knowledge and critical gaps. Global Climate Change and Agricultural Production. 1996:319–30.
-
- Deak MD, Porter WP, Mathewson PD, Lovelace DM, Flores RJ, Tripati AK, et al.. Metabolic skinflint or spendthrift? Insights into ground sloth integument and thermophysiology revealed by biophysical modeling and clumped isotope paleothermometry. J Mamm Evol. 2025;32(1):1. doi: 10.1007/s10914-024-09743-2 - DOI - PMC - PubMed
-
- Acosta-Motos JR, Ortuño MF, Bernal-Vicente A, Diaz-Vivancos P, Sanchez-Blanco M, Hernandez J. Plant Responses to Salt Stress: Adaptive Mechanisms. Agronomy. 2017;7(1):18. doi: 10.3390/agronomy7010018 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

