Modeling diffusive search by non-adaptive sperm: Empirical and computational insights
- PMID: 40244975
- PMCID: PMC12005489
- DOI: 10.1371/journal.pcbi.1012865
Modeling diffusive search by non-adaptive sperm: Empirical and computational insights
Abstract
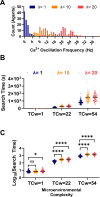
During fertilization, mammalian sperm undergo a winnowing selection process that reduces the candidate pool of potential fertilizers from ~106-1011 cells to 101-102 cells (depending on the species). Classical sperm competition theory addresses the positive or 'stabilizing' selection acting on sperm phenotypes within populations of organisms but does not strictly address the developmental consequences of sperm traits among individual organisms that are under purifying selection during fertilization. It is the latter that is of utmost concern for improving assisted reproductive technologies (ART) because low-fitness sperm may be inadvertently used for fertilization during interventions that rely heavily on artificial sperm selection, such as intracytoplasmic sperm injection (ICSI). Importantly, some form of sperm selection is used in nearly all forms of ART (e.g., differential centrifugation, swim-up, or hyaluronan binding assays, etc.). To date, there is no unifying quantitative framework (i.e., theory of sperm selection) that synthesizes causal mechanisms of selection with observed natural variation in individual sperm traits. In this report, we reframe the physiological function of sperm as a collective diffusive search process and develop multi-scale computational models to explore the causal dynamics that constrain sperm fitness during fertilization. Several experimentally useful concepts are developed, including a probabilistic measure of sperm fitness as well as an information theoretic measure of the magnitude of sperm selection, each of which are assessed under systematic increases in microenvironmental selective pressure acting on sperm motility patterns.
Copyright: © 2025 Brisard et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures



Update of
-
Modeling Diffusive Search by Non-Adaptive Sperm: Empirical and Computational Insights.bioRxiv [Preprint]. 2024 Dec 30:2024.06.17.599386. doi: 10.1101/2024.06.17.599386. bioRxiv. 2024. Update in: PLoS Comput Biol. 2025 Apr 17;21(4):e1012865. doi: 10.1371/journal.pcbi.1012865. PMID: 38948799 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

