The length of the G1 phase is an essential determinant of H3K27me3 landscapes across diverse cell types
- PMID: 40245079
- PMCID: PMC12052206
- DOI: 10.1371/journal.pbio.3003119
The length of the G1 phase is an essential determinant of H3K27me3 landscapes across diverse cell types
Abstract
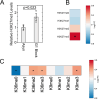
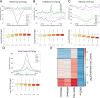
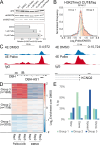
Stem cells have lower facultative heterochromatin as defined by trimethylation of histone H3 lysine 27 (H3K27me3) compared to differentiated cells. However, the mechanisms underlying these differential H3K27me3 levels remain elusive. Because H3K27me3 levels are diluted 2-fold in every round of replication and then restored through the rest of the cell cycle, we reasoned that the cell cycle length could be a key regulator of total H3K27me3 levels. Here, we propose that a short G1 phase restricts H3K27me3 levels in stem cells. To test this model, we determined changes to H3K27me3 levels in mouse embryonic stem cells (mESCs) globally and at specific loci upon G1 phase lengthening - accomplished by thymidine block or growth in the absence of serum (with the "2i medium"). H3K27me3 levels in mESCs increase with G1 arrest when grown in serum and in 2i medium. Additionally, we observed via CUT&RUN and ChIP-seq that regions that gain H3K27me3 in G1 arrest and 2i media overlap, supporting our model of G1 length as a critical regulator of the stem cell epigenome. Furthermore, we demonstrate the inverse effect - that G1 shortening in differentiated human HEK293 cells results in a loss of H3K27me3 levels. Finally, in human tumor cells with extreme H3K27me3 loss, lengthening of the G1 phase leads to H3K27me3 recovery despite the presence of the dominant negative, sub-stoichiometric H3K27M mutation. Our results indicate that G1 length is an essential determinant of H3K27me3 landscapes across diverse cell types.
Copyright: © 2025 Trouth et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Update of
-
G1 length dictates H3K27me3 landscapes.bioRxiv [Preprint]. 2024 Aug 26:2023.12.05.570186. doi: 10.1101/2023.12.05.570186. bioRxiv. 2024. Update in: PLoS Biol. 2025 Apr 17;23(4):e3003119. doi: 10.1371/journal.pbio.3003119. PMID: 38106207 Free PMC article. Updated. Preprint.
Similar articles
-
G1 length dictates H3K27me3 landscapes.bioRxiv [Preprint]. 2024 Aug 26:2023.12.05.570186. doi: 10.1101/2023.12.05.570186. bioRxiv. 2024. Update in: PLoS Biol. 2025 Apr 17;23(4):e3003119. doi: 10.1371/journal.pbio.3003119. PMID: 38106207 Free PMC article. Updated. Preprint.
-
H3K27me3 and the PRC1-H2AK119ub pathway cooperatively maintain heterochromatin and transcriptional silencing after the loss of H3K9 methylation.Epigenetics Chromatin. 2025 May 2;18(1):26. doi: 10.1186/s13072-025-00589-3. Epigenetics Chromatin. 2025. PMID: 40312364 Free PMC article.
-
Contrasting epigenetic states of heterochromatin in the different types of mouse pluripotent stem cells.Sci Rep. 2018 Apr 10;8(1):5776. doi: 10.1038/s41598-018-23822-4. Sci Rep. 2018. PMID: 29636490 Free PMC article.
-
Bivalent histone modifications in early embryogenesis.Curr Opin Cell Biol. 2012 Jun;24(3):374-86. doi: 10.1016/j.ceb.2012.03.009. Epub 2012 Apr 17. Curr Opin Cell Biol. 2012. PMID: 22513113 Free PMC article. Review.
-
JMJD3: a critical epigenetic regulator in stem cell fate.Cell Commun Signal. 2021 Jul 3;19(1):72. doi: 10.1186/s12964-021-00753-8. Cell Commun Signal. 2021. PMID: 34217316 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

