Pleural fluid proteomics from patients with pleural infection shows signatures of diverse neutrophilic responses: The Oxford Pleural Infection Endotyping Study (TORPIDS-2)
- PMID: 40246538
- PMCID: PMC12256804
- DOI: 10.1183/13993003.00010-2025
Pleural fluid proteomics from patients with pleural infection shows signatures of diverse neutrophilic responses: The Oxford Pleural Infection Endotyping Study (TORPIDS-2)
Abstract
Background: Pleural infection is a complex disease with poor clinical outcomes and increasing incidence worldwide, yet its biological endotypes remain unknown.
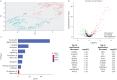
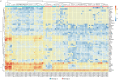
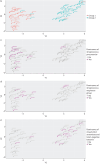
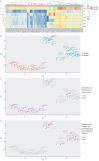
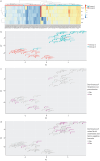
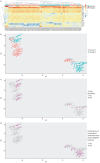
Methods: We analysed 80 pleural fluid samples from the PILOT study, a prospective study on pleural infection, using unlabelled mass spectrometry. A total of 449 proteins were retained after filtering. Unsupervised hierarchical clustering and Uniform Manifold Approximation and Projection analyses were used to cluster samples and pathway analysis was performed to identify the biological processes. Protein signatures as identified by the pathway analysis were compared to microbiology as defined by 16S rRNA next-generation sequencing. Spearman and exact Fischer's methods were used for correlation assessment.
Results: Higher neutrophil degranulation was correlated with increased glycolysis (odds ratio (OR) 281, p<2.2×10-16) and pentose phosphate activation (OR 371.45, p<2.2×10-16). Samples dominated by Streptococcus pneumoniae exhibited higher neutrophil degranulation (OR 12.08, p=0.005), glycolysis (OR 11.4, p=0.006) and pentose phosphate activity (OR 12.82, p=0.004). Samples dominated by anaerobes and Gram-negative bacteria exhibited lower neutrophil degranulation (OR 0.15, p=0.01), glycolysis (OR 0.14, p=0.01) and pentose phosphate activity (OR 0.07, p=0.001). Increased activity of the liver and retinoid X receptors pathway was associated with lower risk of 1-year mortality (OR 0.24, p=0.04).
Conclusions: These findings suggest that pleural infection patients exhibit diverse responses of neutrophil-mediated immunity, glycolysis and pentose phosphate activation, which are associated with microbiology. Therapeutic targeting of the liver and retinoid X receptors pathway with agonists is a possible treatment approach.
Copyright ©The authors 2025.
Conflict of interest statement
Conflict of interest: The authors have no potential conflicts of interest to disclose.
Figures

Comment in
-
Neutrophils in pleural infection: untapped potential for targeted therapeutics.Eur Respir J. 2025 Jul 14;66(1):2500861. doi: 10.1183/13993003.00861-2025. Print 2025 Jul. Eur Respir J. 2025. PMID: 40659465 No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
