Advancing yeast metabolism for a sustainable single carbon bioeconomy
- PMID: 40246696
- PMCID: PMC12020471
- DOI: 10.1093/femsyr/foaf020
Advancing yeast metabolism for a sustainable single carbon bioeconomy
Abstract
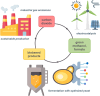
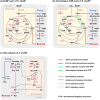
Single carbon (C1) molecules are considered as valuable substrates for biotechnology, as they serve as intermediates of carbon dioxide recycling, and enable bio-based production of a plethora of substances of our daily use without relying on agricultural plant production. Yeasts are valuable chassis organisms for biotech production, and they are able to use C1 substrates either natively or as synthetic engineered strains. This minireview highlights native yeast pathways for methanol and formate assimilation, their engineering, and the realization of heterologous C1 pathways including CO2, in different yeast species. Key features determining the choice among C1 substrates are discussed, including their chemical nature and specifics of their assimilation, their availability, purity, and concentration as raw materials, as well as features of the products to be made from them.
Keywords: bioeconomy; carbon dioxide; formate; methanol; sustainability.
© The Author(s) 2025. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures

References
-
- Bassham JA, Calvin M. The path of carbon in photosynthesis. In PirsonA (ed.), Die CO2-Assimilation/the Assimilation of Carbon Dioxide. Handbuch der Pflanzenphysiologie / Encyclopedia of Plant Physiology. Berlin, Heidelberg: Springer,1960, 884–922.
-
- Berg IA, Kockelkorn D, Buckel W et al. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

