Disease-specific B cell clones are shared between patients with Crohn's disease
- PMID: 40246842
- PMCID: PMC12006383
- DOI: 10.1038/s41467-025-58977-y
Disease-specific B cell clones are shared between patients with Crohn's disease
Abstract
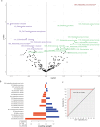
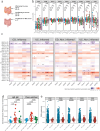
B cells have important functions in gut homeostasis, and dysregulated B cell populations are frequently observed in patients with inflammatory bowel diseases, including both ulcerative colitis (UC) and Crohn's disease (CD). How these B cell perturbations contribute to disease remains largely unknown. Here, we perform deep sequencing of the B cell receptor (BCR) repertoire in four cohorts of patients with CD, together with healthy controls and patients with UC. We identify BCR clones that are shared between patients with CD but not found in healthy individuals nor in patients with UC, indicating CD-associated B cell immune responses. Shared clones are present in the inflamed gut mucosa, draining intestinal lymph nodes and blood, suggesting the presence of common CD-associated antigens that drive B cell responses in CD patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Moor, K. et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature544, 498–502 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

