MIRO1 mutation leads to metabolic maladaptation resulting in Parkinson's disease-associated dopaminergic neuron loss
- PMID: 40246848
- PMCID: PMC12006346
- DOI: 10.1038/s41540-025-00509-x
MIRO1 mutation leads to metabolic maladaptation resulting in Parkinson's disease-associated dopaminergic neuron loss
Abstract
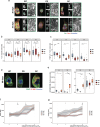
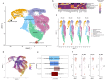
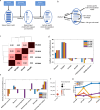
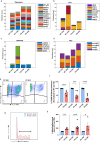
MIRO1 is a mitochondrial outer membrane protein important for mitochondrial distribution, dynamics and bioenergetics. Over the last decade, evidence has pointed to a link between MIRO1 and Parkinson's disease (PD) pathogenesis. Moreover, a heterozygous MIRO1 mutation (p.R272Q) was identified in a PD patient, from which an iPSC-derived midbrain organoid model was derived, showing MIRO1 mutant-dependent selective loss of dopaminergic neurons. Herein, we use patient-specific iPSC-derived midbrain organoids carrying the MIRO1 p.R272Q mutation to further explore the cellular and molecular mechanisms involved in dopaminergic neuron degeneration. Using single-cell RNA sequencing (scRNAseq) analysis and metabolic modeling we show that the MIRO1 p.R272Q mutation affects the dopaminergic neuron developmental path leading to metabolic deficits and disrupted neuron-astrocyte metabolic crosstalk, which might represent an important pathogenic mechanism leading to their loss.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.C.S. is a co-inventor on a patent covering the generation of the here-described midbrain organoids (WO2017060884A1). Furthermore, J.C.S. is a co-founder and shareholder of the company OrganoTherapeutics which makes use of midbrain organoid technology. All other authors declare no competing interests.
Figures

References
-
- Przedborski, S. The two-century journey of Parkinson disease research. Nat. Rev. Neurosci.18, 251–259 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

