An oncohistone-driven H3.3K27M/CREB5/ID1 axis maintains the stemness and malignancy of diffuse intrinsic pontine glioma
- PMID: 40246858
- PMCID: PMC12006333
- DOI: 10.1038/s41467-025-58795-2
An oncohistone-driven H3.3K27M/CREB5/ID1 axis maintains the stemness and malignancy of diffuse intrinsic pontine glioma
Abstract
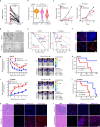
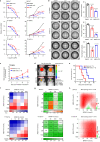
Diffuse intrinsic pontine glioma (DIPG), a lethal pediatric cancer driven by H3K27M oncohistones, exhibits aberrant epigenetic regulation and stem-like cell states. Here, we uncover an axis involving H3.3K27M oncohistones, CREB5/ID1, which sustains the stem-like state of DIPG cells, promoting malignancy. We demonstrate that CREB5 mediates elevated ID1 levels in the H3.3K27M/ACVR1WT subtype, promoting tumor growth; while BMP signaling regulates this process in the H3.1K27M/ACVR1MUT subtype. Furthermore, we reveal that H3.3K27M directly enhances CREB5 expression by reshaping the H3K27me3 landscape at the CREB5 locus, particularly at super-enhancer regions. Additionally, we elucidate the collaboration between CREB5 and BRG1, the SWI/SNF chromatin remodeling complex catalytic subunit, in driving oncogenic transcriptional changes in H3.3K27M DIPG. Intriguingly, disrupting CREB5 super-enhancers with ABBV-075 significantly reduces its expression and inhibits H3.3K27M DIPG tumor growth. Combined treatment with ABBV-075 and a BRG1 inhibitor presents a promising therapeutic strategy for clinical translation in H3.3K27M DIPG treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Anastas, J. N. et al. Re-programing Chromatin with a Bifunctional LSD1/HDAC Inhibitor Induces Therapeutic Differentiation in DIPG. Cancer Cell36, 528–544.e510 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

