Induced proximity to PML protects TDP-43 from aggregation via SUMO-ubiquitin networks
- PMID: 40246979
- PMCID: PMC12394070
- DOI: 10.1038/s41589-025-01886-4
Induced proximity to PML protects TDP-43 from aggregation via SUMO-ubiquitin networks
Abstract
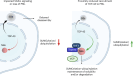
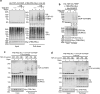
The established role of cytosolic and nuclear inclusions of TDP-43 in the pathogenesis of neurodegenerative disorders has multiplied efforts to understand mechanisms that control TDP-43 aggregation and has spurred searches for approaches limiting this process. Formation and clearance of TDP-43 aggregates are controlled by an intricate interplay of cellular proteostasis systems that involve post-translational modifications and frequently rely on spatial control. We demonstrate that attachment of the ubiquitin-like SUMO2 modifier compartmentalizes TDP-43 in promyelocytic leukemia protein (PML) nuclear bodies and limits the aggregation of TDP-43 in response to proteotoxic stress. Exploiting this pathway through proximity-inducing recruitment of TDP-43 to PML triggers a SUMOylation-ubiquitylation cascade protecting TDP-43 from stress-induced insolubility. The protective function of PML is mediated by ubiquitylation in conjunction with the p97 disaggregase. Altogether, we demonstrate that SUMO-ubiquitin networks protect cells from insoluble TDP-43 inclusions and propose the functionalization of PML as a potential future therapeutic avenue countering aggregation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: E.W. and B.A. have filed a patent on the rapamycin-induced recruitment system used in this work (EP23173977.2). The other authors declare no competing interests. Inclusion and ethics statement: All authors of this study have fulfilled the criteria for authorship required by Nature Portfolio journals.
Figures












References
-
- Hipp, M. S., Kasturi, P. & Hartl, F. U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol.20, 421–435 (2019). - PubMed
-
- Tziortzouda, P., Van Den Bosch, L. & Hirth, F. Triad of TDP43 control in neurodegeneration: autoregulation, localization and aggregation. Nat. Rev. Neurosci.22, 197–208 (2021). - PubMed
-
- Bentmann, E., Haass, C. & Dormann, D. Stress granules in neurodegeneration—lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J.280, 4348–4370 (2013). - PubMed
MeSH terms
Substances
Grants and funding
- Project ID-465470262/Deutsche Forschungsgemeinschaft (German Research Foundation)
- Project ID- 494535244/Deutsche Forschungsgemeinschaft (German Research Foundation)
- CRC387/Deutsche Forschungsgemeinschaft (German Research Foundation)
- project number 442698351/Deutsche Forschungsgemeinschaft (German Research Foundation)
- WO 2108/2-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- Enable/Deutsche Forschungsgemeinschaft (German Research Foundation)Hessisches Ministerium für Wissenschaft und Kunst (Hessen State Ministry of Higher Education, Research and the Arts)
- PROTAC-PDAC:101087045/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- TACTIC/Deutsche Krebshilfe (German Cancer Aid)
LinkOut - more resources
Full Text Sources

