Oxidation of human mitochondrial RNA strongly potentiates immunostimulation in an interferon-associated manner
- PMID: 40247667
- PMCID: PMC12010657
- DOI: 10.1080/13510002.2025.2491845
Oxidation of human mitochondrial RNA strongly potentiates immunostimulation in an interferon-associated manner
Abstract
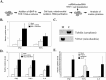
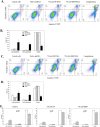
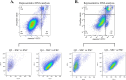
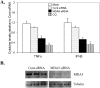
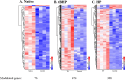
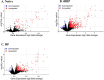
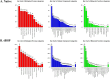
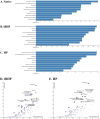
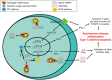
Inflammation is associated with a wide range of medical conditions, most leading causes of death, and high healthcare costs. It can thus benefit from new insights. Here we extended previous studies and found that oxidation of human native mtRNA to 'mitoxRNA' strongly potentiated IFNβ and TNFα immunostimulation in human cells, and that this newly identified type 1 interferon potentiation was transcriptional. This potentiation was significantly greater than with mtDNA oxidation, and t-butylhydroperoxide (tBHP) oxidation of RNA was more proinflammatory than hydrogen peroxide (HP). mtRNA triggered a modest increase in apoptosis that was not potentiated by oxidation, and mtDNA triggered a much greater increase. For native mtRNA, we found that chloroquine-inhibitable endosomes and MDA5 are key signaling pathways for IFNβ and TNFα production. For mitoxRNAs, RNAseq revealed a major increase in both tBHP- and HP-mitoxRNA modulated genes compared with native mtRNA. This increase was very prominent for interferon-related genes, identifying them as important mediators of this powerful oxidation effect. Moderately different gene modulations and KEGG pathways were observed for tBHP- versus HP-mitoxRNAs. These studies reveal the profound effect that mitochondrial RNA oxidation has on immunostimulation, providing new insights into DAMP inflammation and identifying potential therapeutic targets to minimize DAMP mtRNA/mitoxRNA-mediated inflammation.
Keywords: Mitochondria; T-butylhydroperoxide; human; hydrogen peroxide; immunostimulation; oxidative stress; proinflammatory cytokines; type 1 interferons.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
Control (Native) and oxidized (DeMP) mitochondrial RNA are proinflammatory regulators in human.Free Radic Biol Med. 2019 Nov 1;143:62-69. doi: 10.1016/j.freeradbiomed.2019.07.019. Epub 2019 Jul 19. Free Radic Biol Med. 2019. PMID: 31330178
-
Oxidized and Original article degraded mitochondrial polynucleotides (DeMPs), especially RNA, are potent immunogenic regulators in primary mouse macrophages.Free Radic Biol Med. 2017 Mar;104:371-379. doi: 10.1016/j.freeradbiomed.2017.02.009. Epub 2017 Feb 4. Free Radic Biol Med. 2017. PMID: 28179110
-
Mitochondrial Nucleic Acid as a Driver of Pathogenic Type I Interferon Induction in Mendelian Disease.Front Immunol. 2021 Aug 26;12:729763. doi: 10.3389/fimmu.2021.729763. eCollection 2021. Front Immunol. 2021. PMID: 34512665 Free PMC article. Review.
-
Tumor necrosis factor alpha modulates airway smooth muscle function via the autocrine action of interferon beta.J Biol Chem. 2003 Dec 12;278(50):50615-23. doi: 10.1074/jbc.M303680200. Epub 2003 Sep 30. J Biol Chem. 2003. PMID: 14519761
-
Human Mitochondrial RNA Processing and Modifications: Overview.Int J Mol Sci. 2021 Jul 27;22(15):7999. doi: 10.3390/ijms22157999. Int J Mol Sci. 2021. PMID: 34360765 Free PMC article. Review.
References
-
- Cifuentes M, Verdejo HE, Castro PF, et al. . Low-grade chronic inflammation: a shared mechanism for chronic diseases. Physiology (Bethesda). 2025;40(1):0. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
