Structural and Mechanistic Insights into Atypical Bacterial Topoisomerase Inhibitors
- PMID: 40248158
- PMCID: PMC12004755
- DOI: 10.1021/acsmedchemlett.5c00060
Structural and Mechanistic Insights into Atypical Bacterial Topoisomerase Inhibitors
Abstract
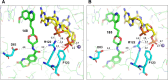
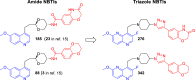
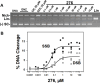
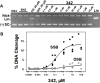
Novel bacterial topoisomerase inhibitors (NBTIs) targeting DNA gyrase and topoisomerase IV constitute a new antibacterial class for deadly pathogens such as MRSA. While most NBTIs induce gyrase-mediated single-strand DNA breaks, a subset of amide NBTIs induces both single-strand and double-strand DNA breaks. Here, we report the X-ray crystal structures of two such amide NBTIs, 148 and 185, and demonstrate an unusual binding mode characterized by engagement of both GyrA D83 and R122. The synthesis of two isosteric triazole NBTIs is also described, one of which (342) affords only single-strand DNA breaks, while the other (276) also induces both single- and double-strand DNA breaks. A combination of docking and molecular dynamics simulations is employed to further investigate the potential structural underpinnings of differences in DNA cleavage.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): M.J.M.-F. is a shareholder of Pfizer.
Figures


References
-
- Mitton-Fry M. J. Novel Bacterial Type II Topoisomerase Inhibitors. Med. Chem. Rev. 2017, 52, 281–302. 10.29200/acsmedchemrev-v52.ch15. - DOI
-
- Bax B. D.; Chan P. F.; Eggleston D. S.; Fosberry A.; Gentry D. R.; Gorrec F.; Giordano I.; Hann M. M.; Hennessy A.; Hibbs M.; Huang J.; Jones E.; Jones J.; Brown K. K.; Lewis C. J.; May E. W.; Saunders M. R.; Singh O.; Spitzfaden C. E.; Shen C.; Shillings A.; Theobald A. J.; Wohlkonig A.; Pearson N. D.; Gwynn M. N. Type IIA Topoisomerase Inhibition by a New Class of Antibacterial Agents. Nature 2010, 466, 935–940. 10.1038/nature09197. - DOI - PubMed
LinkOut - more resources
Full Text Sources

