Rab11b is necessary for mitochondrial integrity and function in gut epithelial cells
- PMID: 40248353
- PMCID: PMC12003269
- DOI: 10.3389/fcell.2025.1498902
Rab11b is necessary for mitochondrial integrity and function in gut epithelial cells
Abstract
Introduction: The RAB11 family of small GTPases are intracellular regulators of membrane and vesicular trafficking. We recently reported that RAB11A and RAB11B redundantly regulate spindle dynamics in dividing gut epithelial cells. However, in contrast to the well-studied RAB11A functions in transporting proteins and lipids through recycling endosomes, the distinct function of RAB11B is less clear.
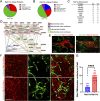
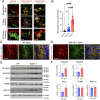
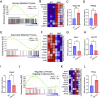
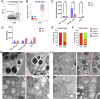
Methods and results: Our proteomic analysis of RAB11A or RAB11B interactome suggested a potential RAB11B specific involvement in regulating mitochondrial functions. Transcriptomic analysis of Rab11b knockout mouse intestines revealed an enhanced mitochondrial protein targeting program with an altered mitochondrial functional integrity. Flow cytometry assessment of mitochondrial membrane potential and reactive oxygen species production revealed an impaired mitochondrial function in vivo. Electron microscopic analysis demonstrated a particularly severe mitochondrial membrane defect in Paneth cells.
Conclusion: These genetic and functional data link RAB11B to mitochondrial structural and functional maintenance for the first time.
Keywords: Paneth cell; Rab11; intestine; mitochondria; proteomics.
Copyright © 2025 Joseph, Han, Bianchi-Smak, Yang, Bhupana, Flores, Delucia, Tran, Goldenring, Bonder and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

