Heparan sulfate proteoglycans remodel SARS-CoV-2 spike conformation to allow integrin interaction and infection of endothelial cells
- PMID: 40248367
- PMCID: PMC12003327
- DOI: 10.3389/fcimb.2025.1552116
Heparan sulfate proteoglycans remodel SARS-CoV-2 spike conformation to allow integrin interaction and infection of endothelial cells
Abstract
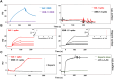
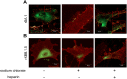
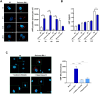
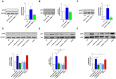
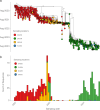
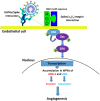
SARS-CoV-2 infects ACE2-negative primary HL-mECs through the interaction of an RGD motif, included in all spike proteins, up to the Omicron BA.1 subvariant, with αvβ3 integrin. Following its entry, SARS-CoV-2 remodels ECs phenotype and promotes angiogenesis in the absence of productive viral replication. Moreover, lack of spike/αvβ3 interaction, occurring in Omicron BA.5 which contains the D405N mutation in the RGD motif, inhibits HL-mECs infection and dysfunction. It is worth noting that anti-spike antibodies do not impact SARS-CoV-2 entry into HL-mECs. This data highlights the fact that i) the RGD motif is not exposed in the entire spike protein and ii) the need of a cofactor favoring spike/αvβ3 interaction. HSPGs are used by different viruses as receptors and coreceptors for their entry into host cells. Here, we use different approaches to scrutinize the role exerted by HSPGs in favoring SARS-CoV-2 infection of ECs. We highlight HSPGs as key molecules responsible for RGD exposure allowing its binding to the αvβ3 integrin as the first step toward viral entry by endocytosis. Indeed, SPR analysis showed lack of spike/αvβ3 interaction in the absence of heparin. This data was further corroborated by immunofluorescence and infectivity assays. Interestingly, the use of Heparinase III or sodium chlorate counteracts the release of proangiogenic molecules and inhibits signaling pathways induced by SARS-CoV-2 infection. Thus, HSPGs may represent a target for preventing SARS-CoV-2 infection of ECs and EC dysfunction-related COVID-19 severity.
Keywords: FAK-Src and Erk signaling pathway; SARS-CoV-2; angiogenesis; heparan sulphates; integrins; von Willebrand factor.
Copyright © 2025 Bugatti, Zani, Bardelli, Giovanetti, Ravelli, Ciccozzi, Caruso and Caccuri.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
References
-
- Bugatti A., Filippini F., Bardelli M., Zani A., Chiodelli P., Messali S., et al. (2022). SARS-CoV-2 infects human ACE2-negative endothelial cells through an integrin-mediated endocytosis even in the presence of vaccine-elicited neutralizing antibodies. Viruses 14, 705. doi: 10.3390/v14040705 - DOI - PMC - PubMed
-
- Bugatti A., Filippini F., Messali S., Giovanetti M., Ravelli C., Zani A., et al. (2023). The D405N Mutation in the spike Protein of SARS-CoV-2 Omicron BA.5 Inhibits spike/Integrins Interaction and Viral Infection of Human Lung Microvascular Endothelial Cells. Viruses 15, 332. doi: 10.3390/v15020332 - DOI - PMC - PubMed
-
- Bugatti A., Marsico S., Mazzuca P., Schulze K., Ebensen T., Giagulli C., et al. (2020). Role of autophagy in von willebrand factor secretion by endothelial cells and in the in vivo thrombin-antithrombin complex formation promoted by the HIV-1 matrix protein p17. Int. J. Mol. Sci. 21, 2022. doi: 10.3390/ijms21062022 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

