Spatial analysis of mitochondrial gene expression reveals dynamic translation hubs and remodeling in stress
- PMID: 40249810
- PMCID: PMC12007585
- DOI: 10.1126/sciadv.ads6830
Spatial analysis of mitochondrial gene expression reveals dynamic translation hubs and remodeling in stress
Abstract
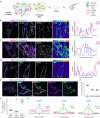
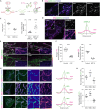
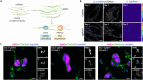
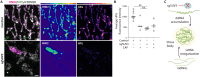
Protein- and RNA-rich bodies contribute to the spatial organization of gene expression in the cell and are also sites of quality control critical to cell fitness. In most eukaryotes, mitochondria harbor their own genome, and all steps of mitochondrial gene expression co-occur within a single compartment-the matrix. Here, we report that processed mitochondrial RNAs are consolidated into micrometer-scale translation hubs distal to mitochondrial DNA transcription and RNA processing sites in human cells. We find that, during stress, mitochondrial messenger and ribosomal RNA are sequestered in mesoscale bodies containing mitoribosome components, concurrent with suppression of active translation. Stress bodies are triggered by proteotoxic stress downstream of double-stranded RNA accumulation in cells lacking unwinding activity of the highly conserved helicase SUPV3L1/SUV3. We propose that the spatial organization of nascent polypeptide synthesis into discrete domains serves to throttle the flow of genetic information to support recovery of mitochondrial quality control.
Figures

Update of
-
A spatial atlas of mitochondrial gene expression reveals dynamic translation hubs and remodeling in stress.bioRxiv [Preprint]. 2024 Aug 17:2024.08.05.604215. doi: 10.1101/2024.08.05.604215. bioRxiv. 2024. Update in: Sci Adv. 2025 Apr 18;11(16):eads6830. doi: 10.1126/sciadv.ads6830. PMID: 39149346 Free PMC article. Updated. Preprint.
References
-
- Suomalainen A., Nunnari J., Mitochondria at the crossroads of health and disease. Cell 187, 2601–2627 (2024). - PubMed
-
- Gustafsson C. M., Falkenberg M., Larsson N.-G., Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 85, 133–160 (2016). - PubMed
-
- Antonicka H., Sasarman F., Nishimura T., Paupe V., Shoubridge E. A., The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 17, 386–398 (2013). - PubMed
-
- Kakudji E. V., Lewis S. C., Mitochondrial nucleoids. Curr. Biol. 34, R1067–R1068 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

