Proteomic-based stemness score measures oncogenic dedifferentiation and enables the identification of druggable targets
- PMID: 40250426
- PMCID: PMC12230239
- DOI: 10.1016/j.xgen.2025.100851
Proteomic-based stemness score measures oncogenic dedifferentiation and enables the identification of druggable targets
Abstract
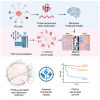
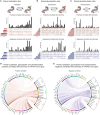
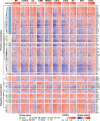
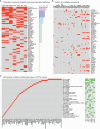
Cancer progression and therapeutic resistance are closely linked to a stemness phenotype. Here, we introduce a protein-expression-based stemness index (PROTsi) to evaluate oncogenic dedifferentiation in relation to histopathology, molecular features, and clinical outcomes. Utilizing datasets from the Clinical Proteomic Tumor Analysis Consortium across 11 tumor types, we validate PROTsi's effectiveness in accurately quantifying stem-like features. Through integration of PROTsi with multi-omics, including protein post-translational modifications, we identify molecular features associated with stemness and proteins that act as active nodes within transcriptional networks, driving tumor aggressiveness. Proteins highly correlated with stemness were identified as potential drug targets, both shared and tumor specific. These stemness-associated proteins demonstrate predictive value for clinical outcomes, as confirmed by immunohistochemistry in multiple samples. The findings emphasize PROTsi's efficacy as a valuable tool for selecting predictive protein targets, a crucial step in customizing anti-cancer therapy and advancing the clinical development of cures for cancer patients.
Keywords: biomarkers; cancer; drug targets; kinase activity; machine learning; mass spectrometry; multiomics; proteomics; stemness; tumor plasticity.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Hanahan D. Hallmarks of cancer: New Dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
-
- Malta T.M., Sokolov A., Gentles A.J., Burzykowski T., Poisson L., Weinstein J.N., Kamińska B., Huelsken J., Omberg L., Gevaert O., et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell. 2018;173:338–354.e15. doi: 10.1016/j.cell.2018.03.034. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

