Selective identification of epigenetic regulators at methylated genomic sites by SelectID
- PMID: 40251151
- PMCID: PMC12008204
- DOI: 10.1038/s41467-025-59002-y
Selective identification of epigenetic regulators at methylated genomic sites by SelectID
Abstract
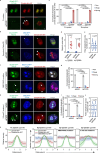
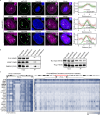
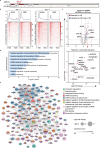
DNA methylation is a significant component in proximal chromatin regulation and plays crucial roles in regulating gene expression and maintaining the repressive state of retrotransposon elements. However, accurate profiling of the proteomics which simultaneously identifies specific DNA sequences and their associated epigenetic modifications remains a challenge. Here, we report a strategy termed SelectID (selective profiling of epigenetic control at genome targets identified by dCas9), which introduces methylated DNA binding domain into dCas9-mediated proximity labeling system to enable in situ protein capture at repetitive elements with 5-methylcytosine (5mC) modifications. SelectID is demonstrated as feasible as dCas9-TurboID system at specific DNA methylation regions, such as the chromosome 9 satellite. Using SelectID, we successfully identify CHD4 as potential repressors of methylated long interspersed nuclear element-1 (LINE-1) retrotransposon through direct binding at the 5' untranslated region (5'UTR) of young LINE-1 elements. Overall, our SelectID approach has opened up avenues for uncovering potential regulators of specific DNA regions with DNA methylation, which will greatly facilitate future studies on epigenetic regulation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5' untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated.Mol Cell Biol. 2007 Jul;27(13):4685-97. doi: 10.1128/MCB.02138-06. Epub 2007 Apr 30. Mol Cell Biol. 2007. PMID: 17470553 Free PMC article.
-
Identification of epigenetically active L1 promoters in the human brain and their relationship with psychiatric disorders.Neurosci Res. 2023 Oct;195:37-51. doi: 10.1016/j.neures.2023.05.001. Epub 2023 May 2. Neurosci Res. 2023. PMID: 37141946
-
Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions.Genome Res. 2009 Aug;19(8):1374-83. doi: 10.1101/gr.089185.108. Epub 2009 Jun 19. Genome Res. 2009. PMID: 19542493 Free PMC article.
-
Alternative roles for oxidized mCs and TETs.Curr Opin Genet Dev. 2017 Feb;42:1-7. doi: 10.1016/j.gde.2016.11.003. Epub 2016 Dec 7. Curr Opin Genet Dev. 2017. PMID: 27939598 Free PMC article. Review.
-
Long Interspersed Element-1 Methylation Level as a Prognostic Biomarker in Gastrointestinal Cancers.Digestion. 2018;97(1):26-30. doi: 10.1159/000484104. Epub 2018 Feb 1. Digestion. 2018. PMID: 29393154 Review.
References
-
- Deniz, O., Frost, J. M. & Branco, M. R. Regulation of transposable elements by DNA modifications (vol 20, pg 417, 2019). Nat. Rev. Genet.20, 432–432 (2019). - PubMed
-
- Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet.13, 484–492 (2012). - PubMed
-
- Smit, A. F., Toth, G., Riggs, A. D. & Jurka, J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol.246, 401–417 (1995). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

