A microengineered 3D human neurovascular unit model to probe the neuropathogenesis of herpes simplex encephalitis
- PMID: 40251168
- PMCID: PMC12008363
- DOI: 10.1038/s41467-025-59042-4
A microengineered 3D human neurovascular unit model to probe the neuropathogenesis of herpes simplex encephalitis
Abstract
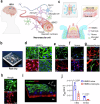
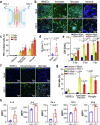
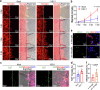
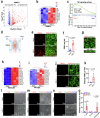
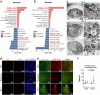
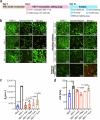
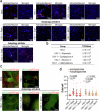
Herpes simplex encephalitis (HSE) caused by HSV-1 is the most common non-epidemic viral encephalitis, and the neuropathogenesis of HSE remains elusive. This work describes a 3D human neurovascular unit (NVU) model that allows to explore the neuropathogenesis of HSE in vitro. This model is established by co-culturing human microvascular endothelial cells, astrocytes, microglia and neurons on a multi-compartment chip. Upon HSV-1 infection, this NVU model exhibited HSE-associated pathological changes, including cytopathic effects, blood-brain barrier dysfunction and pro-inflammatory cytokines release. Besides, significant innate immune responses were observed with the infiltration of peripheral immune cells and microglial activation. Transcriptomic analysis revealed broadly inflammatory and chemotactic responses in host cells. Mechanistically, we found HSV-1 could induce severe suppression of autophagic flux in glial cells, especially in microglia. Autophagy activators could effectively inhibit HSV-1 replication and rescue neurovascular injuries, indicating the utility of this unique platform for studying neurological diseases and new therapeutics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Microglia Activate Early Antiviral Responses upon Herpes Simplex Virus 1 Entry into the Brain to Counteract Development of Encephalitis-Like Disease in Mice.J Virol. 2022 Mar 23;96(6):e0131121. doi: 10.1128/JVI.01311-21. Epub 2022 Jan 19. J Virol. 2022. PMID: 35045263 Free PMC article.
-
Modulation of experimental herpes encephalitis-associated neurotoxicity through sulforaphane treatment.PLoS One. 2012;7(4):e36216. doi: 10.1371/journal.pone.0036216. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558388 Free PMC article.
-
Enhanced viral clearance and reduced leukocyte infiltration in experimental herpes encephalitis after intranasal infection of CXCR3-deficient mice.J Neurovirol. 2017 Jun;23(3):394-403. doi: 10.1007/s13365-016-0508-6. Epub 2017 Jan 23. J Neurovirol. 2017. PMID: 28116674
-
Mechanisms of Blood-Brain Barrier Disruption in Herpes Simplex Encephalitis.J Neuroimmune Pharmacol. 2019 Jun;14(2):157-172. doi: 10.1007/s11481-018-9821-6. Epub 2018 Nov 19. J Neuroimmune Pharmacol. 2019. PMID: 30456443 Review.
-
Microglial response to viral challenges: every silver lining comes with a cloud.Front Biosci (Landmark Ed). 2011 Jun 1;16(6):2187-205. doi: 10.2741/3847. Front Biosci (Landmark Ed). 2011. PMID: 21622170 Review.
References
-
- Marcocci, M. E. et al. Herpes simplex virus-1 in the brain: the dark side of a sneaky infection. Trends Microbiol.28, 808–820 (2020). - PubMed
-
- Stahl, J. P. & Mailles, A. Herpes simplex virus encephalitis update. Curr. Opin. Infect. Dis.32, 239–243 (2019). - PubMed
-
- Gnann, J. W. Jr. & Whitley, R. J. Herpes simplex encephalitis: an update. Curr. Infect. Dis. Rep.19, 13 (2017). - PubMed
-
- Liu, H. et al. Proteomics analysis of HSV-1-induced alterations in mouse brain microvascular endothelial cells. J. Neurovirol.25, 525–539 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

