Safety and feasibility of 4-1BB co-stimulated CD19-specific CAR-NK cell therapy in refractory/relapsed large B cell lymphoma: a phase 1 trial
- PMID: 40251398
- PMCID: PMC12122374
- DOI: 10.1038/s43018-025-00940-3
Safety and feasibility of 4-1BB co-stimulated CD19-specific CAR-NK cell therapy in refractory/relapsed large B cell lymphoma: a phase 1 trial
Erratum in
-
Author Correction: Safety and feasibility of 4-1BB co-stimulated CD19-specific CAR-NK cell therapy in refractory/relapsed large B cell lymphoma: a phase 1 trial.Nat Cancer. 2025 Jul;6(7):1295. doi: 10.1038/s43018-025-01026-w. Nat Cancer. 2025. PMID: 40610723 Free PMC article. No abstract available.
Abstract
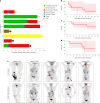
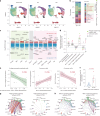
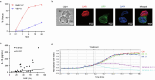
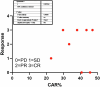
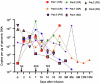
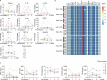
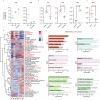
Chimeric antigen receptor (CAR)-modified NK (CAR-NK) cells are candidates for next-generation cancer immunotherapies. Here we generated CD19-specific CAR-NK cells with 4-1BB and CD3ζ signaling endo-domains (CD19-BBz CAR-NK) by transduction of cord blood-derived NK cells using baboon envelope pseudotyped lentiviral vectors and demonstrated their antitumor activity in preclinical B cell lymphoma models in female mice. We next conducted a phase 1 dose-escalation trial involving repetitive administration of CAR-NK cells in 8 patients with relapsed/refractory large B cell lymphoma (NCT05472558). Primary end points were safety, maximum tolerated dose, and overall response rate. Secondary end points included duration of response, overall survival, and progression-free survival. No dose-limiting toxicities occurred, and the maximum tolerated dose was not reached. No cases of cytokine release syndrome, neurotoxicity, or graft-versus-host disease were observed. Results showed an overall response rate of 62.5% at day 30, with 4 patients (50%) achieving complete response. The median progression-free survival was 9.5 months, and the median overall survival was not reached. A post hoc exploratory single-cell RNA sequencing analysis revealed molecular features of CAR-NK cells associated with therapeutic efficacy and efficacy-related immune cell interaction networks. This study met the pre-specified end points. In conclusion, CD19-BBz CAR-NK cells were feasible and therapeutically safe, capable of inducing durable response in patients with B cell lymphoma.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Chong, E. A., Ruella, M., Schuster, S. J. & Lymphoma Program Investigators at the University of Pennsylvania Five-year outcomes for refractory B-cell lymphomas with CAR T-cell therapy. N. Engl. J. Med.384, 673–674 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 82170219/National Natural Science Foundation of China (National Science Foundation of China)
- 2021C03117/Science and Technology Department of Zhejiang Province
- Y24H080001/Natural Science Foundation of Zhejiang Province (Zhejiang Provincial Natural Science Foundation)
- LY23H080004/Natural Science Foundation of Zhejiang Province (Zhejiang Provincial Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

