Prevalence of genes encoding carbapenem-resistance in Klebsiella pneumoniae recovered from clinical samples in Africa: systematic review and meta-analysis
- PMID: 40251495
- PMCID: PMC12007206
- DOI: 10.1186/s12879-025-10959-7
Prevalence of genes encoding carbapenem-resistance in Klebsiella pneumoniae recovered from clinical samples in Africa: systematic review and meta-analysis
Abstract
Background: The potential of Klebsiella pneumoniae (K. pneumoniae) to acquire and spread carbapenem-resistant genes is the most concerning characteristic of the bacteria. In hospitals and other healthcare settings, multidrug-resistant K. pneumoniae can be prevalent and cause severe infections, posing significant challenges to patient management. Studying genetic variants and drug-resistant mutations in pathogenic bacteria of public health importance is essential. Therefore, this study aimed to assess the overall prevalence of carbapenemase-encoding genes in K. pneumoniae across Africa.
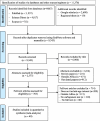
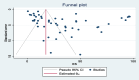
Methods: All studies published between January 2010, and December 2023, were retrieved from the electronic databases PubMed, Science Direct, and Scopus, as well as through the Google Scholar search engine. This systematic review and meta-analysis adhered strictly to the PRISMA guidelines. Data analysis was performed using STATA version 17. The quality of the included studies was critically evaluated using the "Joanna Briggs Institute" criteria. To evaluate heterogeneity among the studies, inverse variance (I2) tests were utilized. Subgroup analysis was conducted when heterogeneity exists among studies. To assess publication bias, we used a funnel plot and Egger's regression test. A random effects model was used to calculate the weighted pooled prevalence of genetic variants associated with carbapenem resistance in K. pneumoniae.
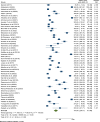
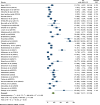
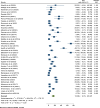
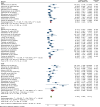
Results: A total of 49 potential studies were included in this systematic review and meta-analysis, encompassing 8,021 K. pneumoniae isolates. Among these isolates, 2,254 (28.1%) carbapenems-resistance-conferring genes were identified. The overall pooled prevalence of carbapenemase-encoding genes in K. pneumoniae isolated from clinical specimens across Africa was found to be 34.0% (95% CI: 26.01-41.98%). Furthermore, the pooled prevalence of the carbapenemase genes blaOXA-48 and blaNDM-1 was 16.96% (95% CI: 12.17-21.76%) and 15.08% (95% CI: 9.79-20.37%), respectively. The pooled prevalence of carbapenemase genes in K. pneumoniae isolates from clinical samples across Africa increased over time, reported as 20.4%(-0.7-41.4%) for 2010-2015, 34.5% (20.2-48.8%) for 2016-2020, and 35.2% (24.8-45.5%) for 2021-2023, with heterogeneity (I2) values of 36.5%, 96.7%, and 99.3%, respectively.
Conclusions: The emergence and spread of carbapenemase-encoding genes in K. pneumoniae pose a major threat to public health. Knowledge on the genetic mechanisms of carbapenem resistance is crucial for developing effective strategies to combat these multidrug-resistant infections and reduce their impact on healthcare systems. The carbapenemase genes blaOXA-48 and blaNDM-1 were the most prevalent and showed an increasing trend over time.
Keywords: Bla NDM-1; Bla OXA-48; K. pneumoniae; Carbapenemase-encoding genes; Clinical samples; Meta-analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not required as it exclusively utilized publicly available aggregated data. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Genomic dynamics of high-risk carbapenem-resistant klebsiella pneumoniae clones carrying hypervirulence determinants in Egyptian clinical settings.BMC Infect Dis. 2024 Oct 22;24(1):1193. doi: 10.1186/s12879-024-10056-1. BMC Infect Dis. 2024. PMID: 39438795 Free PMC article.
-
Prevalence of carbapenem-resistant gram-negative bacteria among neonates suspected for sepsis in Africa: a systematic review and meta-analysis.BMC Infect Dis. 2024 Aug 18;24(1):838. doi: 10.1186/s12879-024-09747-6. BMC Infect Dis. 2024. PMID: 39155370 Free PMC article.
-
Systematic review and meta-analysis on the carbapenem-resistant hypervirulent Klebsiella pneumoniae isolates.BMC Pharmacol Toxicol. 2025 Jan 30;26(1):25. doi: 10.1186/s40360-025-00857-8. BMC Pharmacol Toxicol. 2025. PMID: 39885589 Free PMC article.
-
Prevalence of carbapenemase genes in Klebsiella pneumoniae and Escherichia coli isolated from intensive care unit patients in Iran: a systematic review (2014-2025).Mol Biol Rep. 2025 Jul 1;52(1):660. doi: 10.1007/s11033-025-10774-y. Mol Biol Rep. 2025. PMID: 40590995 Review.
-
Molecular epidemiology of carbapenemase-producing Klebsiella pneumoniae in Gauteng South Africa.Sci Rep. 2024 Nov 9;14(1):27337. doi: 10.1038/s41598-024-70910-9. Sci Rep. 2024. PMID: 39521758 Free PMC article.
Cited by
-
The Microbiological Characteristics and Genomic Surveillance of Carbapenem-Resistant Klebsiella pneumoniae Isolated from Clinical Samples.Microorganisms. 2025 Jul 4;13(7):1577. doi: 10.3390/microorganisms13071577. Microorganisms. 2025. PMID: 40732086 Free PMC article.
References
-
- Hansen DS, Gottschau A, Kolmos HJ. Epidemiology of Klebsiella bacteraemia: a case control study using Escherichia coli bacteraemia as control. J Hosp Infect. 1998;38(2):119–32. - PubMed
-
- World Health Organization. WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance to guide research. Development and strategies to prevent and control antimicrobial resistance. 2024. - PubMed
-
- Knapp KM, English BK. Carbapenems. In Seminars in Pediatric Infectious Diseases. 2001;12(3)175–85. 10.1053/spid.2001.24093.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

