Genome-wide association mapping dissects the selective breeding of determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris L.)
- PMID: 40251997
- PMCID: PMC12135006
- DOI: 10.1093/g3journal/jkaf090
Genome-wide association mapping dissects the selective breeding of determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris L.)
Erratum in
-
Correction to: Genome-wide association mapping dissects the selective breeding of determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris L.).G3 (Bethesda). 2025 Aug 6;15(8):jkaf141. doi: 10.1093/g3journal/jkaf141. G3 (Bethesda). 2025. PMID: 40553442 Free PMC article. No abstract available.
Abstract
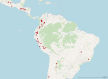
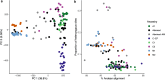
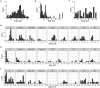
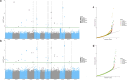
The common bean (Phaseolus vulgaris L.) is a legume pulse crop that provides significant dietary and ecosystem benefits globally. We investigated 2 key traits, determinacy and photoperiod sensitivity, that are integral to its management and crop production, and that were early selected during the domestication of both Mesoamerican and Andean gene pools. Still, significant variation exists among common bean landraces for these traits. Since landraces form the basis for trait introgression in prebreeding, understanding these traits' genetic underpinnings and relation with population structure is vital for guiding breeding and genetic studies. We explored genetic admixture, principal component, and phylogenetic analyses using whole-genome sequencing to define subpopulations and gene pools. We used genome-wide association mapping (GWAS) to identify marker-trait associations in a diversity panel of common bean landraces. We observed a clear correlation between these traits, gene pool, and subpopulation structure. We found extensive admixture between the Andean and Mesoamerican gene pools in some regions. We identified 13 QTLs for determinacy and 10 QTLs for photoperiod sensitivity and underlying causative genes. Our study identified known and novel causative genes and a high proportion of pleiotropic effects for these traits in common bean, and likely translatable to other legume species.
Keywords: GWAS; Plant genetics and genomics; common bean; determinacy; domestication; legume; photoperiod.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflict of interest.
Figures



References
-
- Ambachew D, Londono JM, Castillo NR, Asfaw A, Blair MW. 2024. Genetic diversity, linkage disequilibrium, and population structure in a common bean reference collection. Agronomy. 14(5):985. doi: 10.3390/agronomy14050985. - DOI
-
- Assefa T, Mahama AA, Brown AV, Cannon EKS, Rubyogo JC, Rao IM, Blair MW, Cannon SB. 2019. A review of breeding objectives, genomic resources, and marker-assisted methods in common bean (phaseolus vulgaris l.). Mol Breed. 39(2):1–23. doi: 10.1007/s11032-018-0920-0. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
