Activation of PERK/eIF2α/ATF4 signaling inhibits ERα expression in breast cancer
- PMID: 40252311
- PMCID: PMC12023901
- DOI: 10.1016/j.neo.2025.101165
Activation of PERK/eIF2α/ATF4 signaling inhibits ERα expression in breast cancer
Abstract
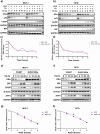
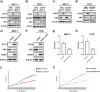
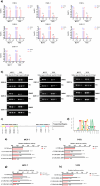
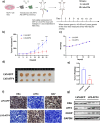
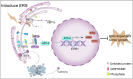
Approximately 70-80% of breast cancers rely on estrogen receptor alpha (ERα) for growth. The unfolded protein response (UPR), a cellular response to endoplasmic reticulum stress (ERS), is an important process crucial for oncogenic transformation. The effect of ERS on ERα expression and signaling remains incompletely elucidated. Here, we focused on the regulatory mechanisms of ERS on ERα expression in ER-positive breast cancer (ER+ BC). Our results demonstrate that ERα protein and mRNA levels in ER+ BC cells are considerably reduced by the ERS inducers thapsigargin (TG) and brefeldin A (BFA) via the PERK/eIF2α/ATF4 signaling pathway. ChIP-qPCR and luciferase reporter gene analysis revealed that ERS induction facilitated ATF4 binding to the ESR1 (the gene encoding ERα) promoter region, thereby suppressing ESR1 promoter activity and inhibiting ERα expression. Furthermore, selective activation of PERK signaling or ATF4 overexpression attenuated ERα expression and tumor cell growth both in vitro and in vivo. In conclusion, our results demonstrate that ERS suppresses ERα expression transcriptionally via the PERK/eIF2α/ATF4 signaling. Our study provides insights into the treatment of ER+ BC by targeting ERα signaling through selective activation of the PERK branch of the UPR.
Keywords: ATF4; Breast cancer; ERα; ESR1; Endoplasmic reticulum stress; Unfolded protein response.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
PARP-1 as a novel target in endocrine-resistant breast cancer.J Exp Clin Cancer Res. 2025 Jun 16;44(1):175. doi: 10.1186/s13046-025-03441-4. J Exp Clin Cancer Res. 2025. PMID: 40518539 Free PMC article.
-
PERK-eIF2α-ATF4 pathway mediated by endoplasmic reticulum stress response is involved in osteodifferentiation of human periodontal ligament cells under cyclic mechanical force.Cell Signal. 2016 Aug;28(8):880-6. doi: 10.1016/j.cellsig.2016.04.003. Epub 2016 Apr 11. Cell Signal. 2016. PMID: 27079961
-
Protein-rich foods, sea foods, and gut microbiota amplify immune responses in chronic diseases and cancers - Targeting PERK as a novel therapeutic strategy for chronic inflammatory diseases, neurodegenerative disorders, and cancer.Pharmacol Ther. 2024 Mar;255:108604. doi: 10.1016/j.pharmthera.2024.108604. Epub 2024 Feb 13. Pharmacol Ther. 2024. PMID: 38360205 Free PMC article. Review.
-
[Cannabidiol inhibits neuronal endoplasmic reticulum stress and apoptosis in rats with multiple concussions by regulating the PERK-eIF2α-ATF4-CHOP pathway].Nan Fang Yi Ke Da Xue Xue Bao. 2025 Jun 20;45(6):1240-1250. doi: 10.12122/j.issn.1673-4254.2025.06.13. Nan Fang Yi Ke Da Xue Xue Bao. 2025. PMID: 40579137 Free PMC article. Chinese.
-
The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress.Curr Mol Med. 2016;16(6):533-44. doi: 10.2174/1566524016666160523143937. Curr Mol Med. 2016. PMID: 27211800 Free PMC article. Review.
References
-
- Engelsman E., Korsten C.B., Persijn J.P., et al. Human breast cancer and estrogen receptor. Arch. Chir. Neerl. 1973;25(4):393–397. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

