Etoposide-induced protein 2.4 homolog promotes argininosuccinate synthase 1 and cancer cell survival upon arginine deprivation
- PMID: 40253325
- PMCID: PMC12008907
- DOI: 10.1186/s11658-025-00726-6
Etoposide-induced protein 2.4 homolog promotes argininosuccinate synthase 1 and cancer cell survival upon arginine deprivation
Abstract
Background: Arginine auxotrophy has been reported in a subset of cancers with inherently defective de novo arginine synthesis. However, the use of arginine deprivation therapy seems to be unequally effective, partially owing to the resistance acquired by cancer cells. Study of underlying factors involved in this response thus becomes of utmost importance. Meanwhile, the function of etoposide-induced 2.4 homolog (EI24) in cancer metabolism, and specifically in arginine metabolism, remains unknown.
Methods: EI24 was overexpressed in cancer cells using a doxycycline-inducible system or adenovirus transduction, while siRNA was used to knockdown EI24. Amino acid(s) deprivation medium was exploited with a cell viability assay to check the reliance of cancer cell survival on arginine. Protein expression and activation were examined through western blot and co-immunoprecipitation blot. Furthermore, global and specific protein translation were assessed through the SUnSET assay and polysome fractionation analysis. Gene expression and arginine level were downloaded from public cancer datasets for in silico validation including gene set enrichment and survival analysis to objectively evaluate the association between EI24 and arginine metabolism.
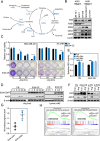
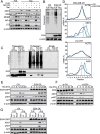
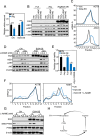
Results: EI24 promoted cancer survival under arginine starvation. Mechanistically, EI24 replenished translation of argininosuccinate synthase 1 (ASS1) by inducing the inactive S-nitrosylated form of phosphatase and tensin homolog (PTEN), leading to release of the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) axis. This tumor-promoting action of EI24 could be found in multiple ASS1-deficient cancer cells regardless of p53 status. Furthermore, expression of EI24 was linked to enrichment of arginine metabolism pathway as well as poor survival of patients with cancer across various cancer types, suggesting its role in cancer resistance to arginine deprivation.
Conclusions: This study is the first to report the role of EI24 in promoting cancer survival via translational regulation of the metabolic enzyme ASS1, thus paving a route for further investigation into the link between EI24 and cancer metabolism.
Keywords: ASS1; Arginine; Cancer metabolism; EI24.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Argininosuccinate synthase 1 is an intrinsic Akt repressor transactivated by p53.Sci Adv. 2017 May 19;3(5):e1603204. doi: 10.1126/sciadv.1603204. eCollection 2017 May. Sci Adv. 2017. PMID: 28560349 Free PMC article.
-
Cisplatin-induced synthetic lethality to arginine-starvation therapy by transcriptional suppression of ASS1 is regulated by DEC1, HIF-1α, and c-Myc transcription network and is independent of ASS1 promoter DNA methylation.Oncotarget. 2016 Dec 13;7(50):82658-82670. doi: 10.18632/oncotarget.12308. Oncotarget. 2016. PMID: 27765932 Free PMC article.
-
Histone deacetylase inhibition is synthetically lethal with arginine deprivation in pancreatic cancers with low argininosuccinate synthetase 1 expression.Theranostics. 2020 Jan 1;10(2):829-840. doi: 10.7150/thno.40195. eCollection 2020. Theranostics. 2020. PMID: 31903153 Free PMC article.
-
Advances in the impact of ASS1 dysregulation on metabolic reprogramming of tumor cells.Cell Signal. 2025 Mar;127:111593. doi: 10.1016/j.cellsig.2025.111593. Epub 2025 Jan 6. Cell Signal. 2025. PMID: 39778698 Review.
-
Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer.Int J Cancer. 2010 Jun 15;126(12):2762-72. doi: 10.1002/ijc.25202. Int J Cancer. 2010. PMID: 20104527 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

