Predicting Nottingham grade in breast cancer digital pathology using a foundation model
- PMID: 40253353
- PMCID: PMC12008962
- DOI: 10.1186/s13058-025-02019-4
Predicting Nottingham grade in breast cancer digital pathology using a foundation model
Erratum in
-
Correction: Predicting Nottingham grade in breast cancer digital pathology using a foundation model.Breast Cancer Res. 2025 May 19;27(1):84. doi: 10.1186/s13058-025-02047-0. Breast Cancer Res. 2025. PMID: 40389997 Free PMC article. No abstract available.
Abstract
Background: The Nottingham histologic grade is crucial for assessing severity and predicting prognosis in breast cancer, a prevalent cancer worldwide. Traditional grading systems rely on subjective expert judgment and require extensive pathological expertise, are time-consuming, and often lead to inter-observer variability.
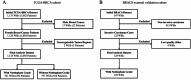
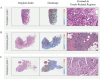
Methods: To address these limitations, we develop an AI-based model to predict Nottingham grade from whole-slide images of hematoxylin and eosin (H&E)-stained breast cancer tissue using a pathology foundation model. From TCGA database, we trained and evaluated using 521 H&E breast cancer slide images with available Nottingham scores through internal split validation, and further validated its clinical utility using an additional set of 597 cases without Nottingham scores. The model leveraged deep features extracted from a pathology foundation model (UNI) and incorporated 14 distinct multiple instance learning (MIL) algorithms.
Results: The best-performing model achieved an F1 score of 0.731 and a multiclass average AUC of 0.835. The top 300 genes correlated with model predictions were significantly enriched in pathways related to cell division and chromosome segregation, supporting the model's biological relevance. The predicted grades demonstrated statistically significant association with 5-year overall survival (p < 0.05).
Conclusion: Our AI-based automated Nottingham grading system provides an efficient and reproducible tool for breast cancer assessment, offering potential for standardization of histologic grade in clinical practice.
Keywords: Biological processes; Breast cancer; Gene expression data; Gene ontology; Multiple instance learning; Nottingham grade; TCGA.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study utilized publicly available and de-identified data from The Cancer Genome Atlas (TCGA) database and the BRACS dataset. Since the data are de-identified and publicly accessible, no additional ethical approval or informed consent was required for this study. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Artificial intelligence grading of breast cancer: a promising method to refine prognostic classification for management precision.Histopathology. 2021 Aug;79(2):187-199. doi: 10.1111/his.14354. Epub 2021 May 6. Histopathology. 2021. PMID: 33590486
-
Multiple instance learning-based prediction of programmed death-ligand 1 (PD-L1) expression from hematoxylin and eosin (H&E)-stained histopathological images in breast cancer.PeerJ. 2025 Apr 15;13:e19201. doi: 10.7717/peerj.19201. eCollection 2025. PeerJ. 2025. PMID: 40256728 Free PMC article.
-
Exploring the Nottingham classification: assessing gene expression profiles in breast cancer patients and their association with outcomes.Breast Cancer Res Treat. 2025 Jul;212(2):237-250. doi: 10.1007/s10549-025-07718-2. Epub 2025 May 28. Breast Cancer Res Treat. 2025. PMID: 40425927
-
Artificial intelligence in digital histopathology for predicting patient prognosis and treatment efficacy in breast cancer.Expert Rev Mol Diagn. 2024 May;24(5):363-377. doi: 10.1080/14737159.2024.2346545. Epub 2024 May 9. Expert Rev Mol Diagn. 2024. PMID: 38655907 Review.
-
Prognostic models for breast cancer: a systematic review.BMC Cancer. 2019 Mar 14;19(1):230. doi: 10.1186/s12885-019-5442-6. BMC Cancer. 2019. PMID: 30871490 Free PMC article.
References
-
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660. - PubMed
-
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–63. 10.3322/caac.21834. - PubMed
-
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–10. 10.1111/j.1365-2559.1991.tb00229.x. - PubMed
-
- Rakha EA, El-Sayed ME, Lee AH, et al. Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J Clin Oncol. 2008;26(19):3153–8. 10.1200/JCO.2007.15.5986. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

