A vasculature-resident innate lymphoid cell population in mouse lungs
- PMID: 40253407
- PMCID: PMC12009297
- DOI: 10.1038/s41467-025-58982-1
A vasculature-resident innate lymphoid cell population in mouse lungs
Abstract
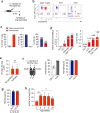
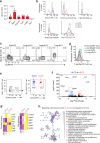
Tissue-resident immune cells such as innate lymphoid cells (ILC) are known to reside in the parenchymal compartments of tissues and modulate local immune protection. Here we use intravascular cell labeling, parabiosis and multiplex 3D imaging to identify a population of group 3 ILCs in mice that are present within the intravascular space of lung blood vessels (vILC3). vILC3s are distributed broadly in alveolar capillary beds from which inhaled pathogens enter the lung parenchyma. By contrast, conventional ILC3s in tissue parenchyma are enriched in lymphoid clusters in proximity to large veins. In a mouse model of pneumonia, Pseudomonas aeruginosa infection results in rapid vILC3 expansion and production of chemokines including CCL4. Blocking CCL4 in vivo attenuates neutrophil recruitment to the lung at the early stage of infection, resulting in prolonged inflammation and delayed bacterial clearance. Our findings thus define the intravascular space as a site of ILC residence in mice, and reveal a unique immune cell population that interfaces with tissue alarmins and the circulating immune system for timely host defense.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
A Mouse Model of Pseudomonas aeruginosa Pneumonia.Methods Mol Biol. 2021;2321:53-61. doi: 10.1007/978-1-0716-1488-4_6. Methods Mol Biol. 2021. PMID: 34048007
-
Candida albicans airway exposure primes the lung innate immune response against Pseudomonas aeruginosa infection through innate lymphoid cell recruitment and interleukin-22-associated mucosal response.Infect Immun. 2014 Jan;82(1):306-15. doi: 10.1128/IAI.01085-13. Epub 2013 Oct 28. Infect Immun. 2014. PMID: 24166952 Free PMC article.
-
Innate immune responses to Pseudomonas aeruginosa infection.Microbes Infect. 2011 Dec;13(14-15):1133-45. doi: 10.1016/j.micinf.2011.07.011. Epub 2011 Aug 2. Microbes Infect. 2011. PMID: 21839853 Free PMC article. Review.
-
Role of Toll-like receptor 5 in the innate immune response to acute P. aeruginosa pneumonia.Am J Physiol Lung Cell Mol Physiol. 2009 Dec;297(6):L1112-9. doi: 10.1152/ajplung.00155.2009. Epub 2009 Oct 2. Am J Physiol Lung Cell Mol Physiol. 2009. PMID: 19801452 Free PMC article.
-
Pseudomonas aeruginosa in chronic lung disease: untangling the dysregulated host immune response.Front Immunol. 2024 Jun 28;15:1405376. doi: 10.3389/fimmu.2024.1405376. eCollection 2024. Front Immunol. 2024. PMID: 39015565 Free PMC article. Review.
References
-
- Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature517, 293–301 (2015). - PubMed
-
- Klose, C. S. & Artis, D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol.17, 765–774 (2016). - PubMed
-
- Juelke, K. & Romagnani, C. Differentiation of human innate lymphoid cells (ILCs). Curr. Opin. Immunol.38, 75–85 (2016). - PubMed
MeSH terms
Grants and funding
- HL159675/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 HL152293/HL/NHLBI NIH HHS/United States
- 1R35GM138805/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- HL152293/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 HL159675/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

