Structural characterization of antibody-responses following Zolgensma treatment for AAV capsid engineering to expand patient cohorts
- PMID: 40253479
- PMCID: PMC12009303
- DOI: 10.1038/s41467-025-59088-4
Structural characterization of antibody-responses following Zolgensma treatment for AAV capsid engineering to expand patient cohorts
Abstract
Monoclonal antibodies are useful tools to dissect the neutralizing antibody response against the adeno-associated virus (AAV) capsids that are used as gene therapy delivery vectors. The presence of pre-existing neutralizing antibodies in large portions of the human population poses a significant challenge for AAV-mediated gene therapy, primarily targeting the capsid leading to vector inactivation and loss of treatment efficacy. This study structurally characterizes the interactions of 21 human-derived neutralizing antibodies from three patients treated with the AAV9 vector, Zolgensma®, utilizing high-resolution cryo-electron microscopy. The antibodies bound to the 2-fold depression or the 3-fold protrusions do not conform to the icosahedral symmetry of the capsid, thus requiring localized reconstructions. These complex structures provide unprecedented details of the mAbs binding interfaces, with many antibodies inducing structural perturbations of the capsid upon binding. Key surface capsid amino acid residues were identified facilitating the design of capsid variants with antibody escape phenotypes. These AAV9 capsid variants have the potential to expand the patient cohort to include those that were previously excluded due to their pre-existing neutralizing antibodies against the wtAAV9 capsid, and the possibly of further treatment to those requiring redosing.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: IAE and GJL are in advanced discussion with a biotechnology company regarding the commercial manufacture and distribution of the monoclonal antibodies (mAbs) described in this manuscript. These negotiations may result in financial compensation for IAE and GJL and their affiliated organizations. JTH is co-founder and CEO of Nanometria, a limited liability company. The University of Florida Research Foundation, Inc. has filed a patent application (PCT/US2024/015006 [ https://patents.google.com/patent/WO2024168153A2/en?oq=PCT%2fUS2024%2f015006 ]) on behalf of MM, JH, ARN, and RM based on the capsid variants described in this study. The remaining authors declare no competing interests.
Figures
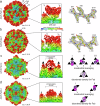




Update of
-
Structural characterization of antibody-responses from Zolgensma treatment provides the blueprint for the engineering of an AAV capsid suitable for redosing.bioRxiv [Preprint]. 2024 May 3:2024.05.01.590489. doi: 10.1101/2024.05.01.590489. bioRxiv. 2024. Update in: Nat Commun. 2025 Apr 19;16(1):3731. doi: 10.1038/s41467-025-59088-4. PMID: 38746165 Free PMC article. Updated. Preprint.
References
-
- Keam, S. J. Eladocagene exuparvovec: first approval. Drugs82, 1427–1432 (2022). - PubMed
MeSH terms
Substances
Grants and funding
- APP2029992/Department of Health | National Health and Medical Research Council (NHMRC)
- U24 GM129541/GM/NIGMS NIH HHS/United States
- R01 GM082946/GM/NIGMS NIH HHS/United States
- APP1194940/Department of Health | National Health and Medical Research Council (NHMRC)
- 348021/Academy of Finland (Suomen Akatemia)
LinkOut - more resources
Full Text Sources

