Drug and siRNA screens identify ROCK2 as a therapeutic target for ciliopathies
- PMID: 40253509
- PMCID: PMC12009310
- DOI: 10.1038/s43856-025-00847-1
Drug and siRNA screens identify ROCK2 as a therapeutic target for ciliopathies
Abstract
Background: Primary cilia mediate vertebrate development and growth factor signalling. Defects in primary cilia cause inherited developmental conditions termed ciliopathies. Ciliopathies often present with cystic kidney disease, a major cause of early renal failure. Currently, only one drug, Tolvaptan, is licensed to slow the decline of renal function for the ciliopathy polycystic kidney disease. Novel therapeutic interventions are needed.
Methods: We screened clinical development compounds to identify those that reversed cilia loss due to siRNA knockdown. In parallel, we undertook a whole genome siRNA-based reverse genetics phenotypic screen to identify positive modulators of cilia formation.
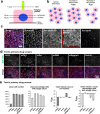
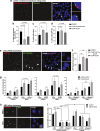
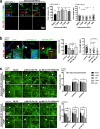
Results: Using a clinical development compound screen, we identify fasudil hydrochloride. Fasudil is a generic, off-patent drug that is a potent, broadly selective Rho-associated coiled-coil-containing protein kinase (ROCK) inhibitor. In parallel, the siRNA screen identifies ROCK2 and we demonstrate that ROCK2 is a key mediator of cilium formation and function through its possible effects on actin cytoskeleton remodelling.
Conclusions: Our results indicate that specific ROCK2 inhibitors (e.g. belumosudil) could be repurposed for cystic kidney disease treatment. We propose that ROCK2 inhibition represents a novel, disease-modifying therapeutic approach for heterogeneous ciliopathies.
Plain language summary
Primary cilia are antennae-like structures on cells that are important for early development and healthy cell function. Defects in primary cilia can cause inherited diseases called ciliopathies. Ciliopathies often cause fluid-filled sacs, called cysts, that are a major cause of kidney disease and failure. There is currently one drug licensed to slow kidney disease progression, but it is poorly tolerated in patients. Therefore, new drugs are needed. In this study, we used screening assays to identify potential drugs and their targets that are effective in promoting the formation of primary cilia. Our results identified ROCK2 (Rho-associated coiled-coil-containing protein kinase 2), an inhibitor of protein signalling, as a key mediator of cilium function. These findings suggest that drugs that specifically target ROCK2 could be a potential treatment option for cystic kidney disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

