GAS-J, a User-Friendly Browser Application for Genome Assembly, emm-Typing, MLST Typing, and Virulence Factor Gene Detection of Streptococcus pyogenes
- PMID: 40253701
- PMCID: PMC12232109
- DOI: 10.1111/1348-0421.13223
GAS-J, a User-Friendly Browser Application for Genome Assembly, emm-Typing, MLST Typing, and Virulence Factor Gene Detection of Streptococcus pyogenes
Abstract
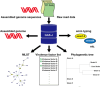
Clinical isolates of Streptococcus pyogenes are usually classified using emm and multilocus sequence typing (MLST). Recently, whole genome sequencing (WGS) has been employed for emm typing and MLST analysis. WGS data provides additional information on the presence of virulence factor genes. To enable researchers unfamiliar with bioinformatics to analyze WGS data of S. pyogenes, we opened an online tool named GAS-J, which automatically outputs emm types, sequence types (STs), carriage of virulence factor genes, and phylogenetic trees. The tool accepts raw short-read data as inputs, since it includes the velvet assembler. In all isolates, the emm typing results from this tool were identical to those obtained by conventional PCR and Sanger sequencing, even in cases where isolates had pseudo-emm (emm-like) genes. STs are determined by performing a BLAST search using seven alleles as references. To detect S. pyogenes virulence factor genes, we prepared a new data set containing 620 related proteins. Users may choose which isolates to include in SNP-based phylogenetic tree from a pool of 406 isolates with epidemiological data. The data set includes isolates whose symptoms (STSS or non-STSS) were diagnosed based on the STSS criteria of the Japan Communicable Disease Prevention Law. GAS-J application is available at http://gasj.ncgm.go.jp. The data of isolates are going to be updated in the future.
Keywords: Streptococcus pyogenes; emm typing; multilocus sequence typing; virulence factor; web tool.
© 2025 The Author(s). Microbiology and Immunology published by The Societies and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflicts of interest. Kohei Ogura is an Editorial Board member and is also a coauthor of this article. To minimize bias, he was excluded from all editorial decision‐making related to the acceptance of this article for publication. Late Tohru Miyoshi‐Akiyama served on the Editorial Board until 2023, but is no longer a member of the Board and is therefore not involved in any editorial decision‐making.
Figures

References
-
- Stevens D. L., “Invasive Group A Streptococcus Infections,” Clinical Infectious Diseases 14 (1992): 2–13. - PubMed
-
- Stevens D. L., Bryant A. E., and Yan S., “Invasive Group A Streptococcal Infection: New Concepts in Antibiotic Treatment,” International Journal of Antimicrobial Agents 4 (1994): 297–301. - PubMed
-
- Lancefield R. C., “Current Knowledge of Type‐Specific M Antigens of Group A Streptococci,” Journal of Immunology 89 (1962): 307–313. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

