Histology-resolved proteomics reveals distinct tumor and stromal profiles in low- and high-grade prostate cancer
- PMID: 40254573
- PMCID: PMC12009531
- DOI: 10.1186/s12014-025-09534-8
Histology-resolved proteomics reveals distinct tumor and stromal profiles in low- and high-grade prostate cancer
Abstract
Background: Prostate cancer is one of the most frequently diagnosed cancers in men. Prostate tumor staging and disease aggressiveness are evaluated based on the Gleason scoring system, which is further used to direct clinical intervention. The Gleason scoring system provides an estimate of tumor aggressiveness through quantitation of the serum level of prostate specific antigen (PSA) and histologic assessment of Grade Group, determined by the Gleason Grade of the tumor specimen.
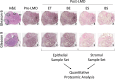
Methods: To improve our understanding of the proteomic characteristics differentiating low- versus high-grade prostate cancer tumors, we performed a deep proteomic characterization of laser microdissected epithelial and stromal subpopulations from surgically resected tissue specimens from patients with Gleason 6 (n = 23 specimens from n = 15 patients) and Gleason 9 (n = 15 specimens from n = 15 patients) prostate cancer via quantitative high-resolution liquid chromatography-tandem mass spectrometry analysis.
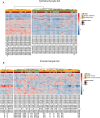
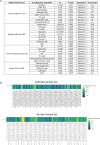
Results: In total, 789 and 295 grade-specific significantly altered proteins were quantified in the tumor epithelium and tumor-involved stroma, respectively. Benign epithelial and stromal populations were not inherently different between Gleason 6 versus Gleason 9 specimens. Notably, 598 proteins were exclusively significantly altered between Gleason 9 (but not Gleason 6) tumor-involved stroma and benign stroma, including several proteins involved in cholesterol biosynthesis and nucleotide metabolism.
Conclusions: Proteomic alterations between Gleason 6 versus Gleason 9 were exclusive to the disease microenvironment, observed in both the tumor epithelium and tumor-involved stroma. Further, the molecular alterations measured in the tumor-involved stroma from Gleason 9 cases relative to the benign stroma have unique significance in disease aggressiveness, development, and/or progression. Our data provide supportive evidence of a need for further investigations into targeting stromal reservoirs of cholesterol and/or deoxynucleoside triphosphates in PCa tumors and further highlight the necessity for independent examination of the TME epithelial and stromal compartments.
Keywords: Laser microdissection; Prostate cancer; Proteomics; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Archival formalin-fixed paraffin embedded (FFPE) tissues were obtained from an IRB-approved protocol (#12–1169, 20122048) from Inova Fairfax Medical Campus (Falls Church, VA, USA). All experimental protocols involving human data in this study were in accordance with the Declaration of Helsinki. Consent for publication: WIRB-Copernicus Group Institutional Review Board (WCG IRB) approved this study. All participants provided written informed consent. The written informed consent included the provision to analyze and publish information and data regarding the results and data from molecular testing, such as proteomics and nucleic acid sequencing. Competing interests: T.P.C. is a ThermoFisher Scientific, Inc. SAB member and receives research funding from AbbVie.
Figures


References
-
- Network NCC. Prostate cancer (version 2.2023). 2023. https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1459.
-
- Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA. The 2014 International society of urological pathology (ISUP) consensus conference on gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol. 2016;40(2):244–52. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
