Anti-SARS-CoV-2 Small Molecule Targeting of Oxysterol-Binding Protein (OSBP) Activates Cellular Antiviral Innate Immunity
- PMID: 40255103
- PMCID: PMC12070403
- DOI: 10.1021/acsinfecdis.4c00631
Anti-SARS-CoV-2 Small Molecule Targeting of Oxysterol-Binding Protein (OSBP) Activates Cellular Antiviral Innate Immunity
Abstract
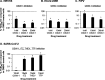
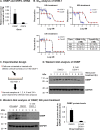
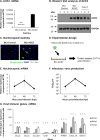
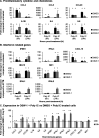
Human oxysterol-binding protein (OSBP) is a potentially druggable mediator in the replication of a broad spectrum of positive-sense (+) single-stranded RNA (ssRNA) viruses, including members of the Picornaviridae, Flaviviridae, and Coronaviridae. OSBP is a cytoplasmic lipid transporting protein capable of moving cholesterol and phosphoinositides between the endoplasmic reticulum (ER) and Golgi, and the ER and lysosome. Several structurally diverse antiviral compounds have been reported to function through targeting OSBP, including the natural product compound OSW-1. Our prior work shows that transient OSW-1 treatment induces a reduction in OSBP protein levels over multiple successive cell generations (i.e., multigenerational), with no apparent cellular toxicity, and the OSW-1-induced reduction of OSBP has antiviral activity against multiple (+)ssRNA viruses. This study extends these findings and establishes that OSW-1 has in vitro antiviral activity against multiple pathogenic (+)ssRNA viruses, including human rhinovirus (HRV1B), the feline coronavirus peritonitis virus (FIPV), human coronavirus 229E (HCoV-229E), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). We also demonstrate that OSW-1 treatment in human airway epithelial cells alters the expression of multiple antiviral innate immune mediators, including the interferon (IFN) related genes IFNB1, IFNL3, CXCL10, ISG15, and MX1. Furthermore, OSW-1 enhances the induction of specific components of type I and III IFN antiviral responses triggered by the RNA viral mimetic polyinosinic-polycytidylic acid (Poly IC). In summary, this study further demonstrates the importance of OSBP in (+)ssRNA virus replication and presents OSBP as a potential regulator of cellular antiviral innate immune responses.
Keywords: OSW-1; SARS-CoV-2; antiviral compounds; innate antiviral immune response; oxysterol-binding protein (OSBP); rhinovirus.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Transient Compound Treatment Induces a Multigenerational Reduction of Oxysterol-Binding Protein (OSBP) Levels and Prophylactic Antiviral Activity.ACS Chem Biol. 2019 Feb 15;14(2):276-287. doi: 10.1021/acschembio.8b00984. Epub 2019 Jan 11. ACS Chem Biol. 2019. PMID: 30576108 Free PMC article.
-
Differing activities of oxysterol-binding protein (OSBP) targeting anti-viral compounds.Antiviral Res. 2019 Oct;170:104548. doi: 10.1016/j.antiviral.2019.104548. Epub 2019 Jul 2. Antiviral Res. 2019. PMID: 31271764 Free PMC article.
-
Oxysterole-binding protein targeted by SARS-CoV-2 viral proteins regulates coronavirus replication.Front Cell Infect Microbiol. 2024 Jul 25;14:1383917. doi: 10.3389/fcimb.2024.1383917. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39119292 Free PMC article.
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
Interferon Control of Human Coronavirus Infection and Viral Evasion: Mechanistic Insights and Implications for Antiviral Drug and Vaccine Development.J Mol Biol. 2022 Mar 30;434(6):167438. doi: 10.1016/j.jmb.2021.167438. Epub 2022 Jan 3. J Mol Biol. 2022. PMID: 34990653 Free PMC article. Review.
References
-
- Roberts B. L.; Severance Z. C.; Bensen R. C.; Le-McClain A. T.; Malinky C. A.; Mettenbrink E. M.; Nuñez J. I.; Reddig W. J.; Blewett E. L.; Burgett A. W. G. Differing Activities of Oxysterol-Binding Protein (OSBP) Targeting Anti-Viral Compounds. Antiviral Res. 2019, 170, 10454810.1016/j.antiviral.2019.104548. - DOI - PMC - PubMed
-
- Roberts B. L.; Severance Z. C.; Bensen R. C.; Le A. T.; Kothapalli N. R.; Nuñez J. I.; Ma H.; Wu S.; Standke S. J.; Yang Z.; Reddig W. J.; Blewett E. L.; Burgett A. W. G. Transient Compound Treatment Induces a Multigenerational Reduction of Oxysterol-Binding Protein (OSBP) Levels and Prophylactic Antiviral Activity. ACS Chem. Biol. 2019, 14 (2), 276–287. 10.1021/acschembio.8b00984. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

